Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters
- PMID: 16352848
- PMCID: PMC1317602
- DOI: 10.1128/JB.188.1.317-327.2006
Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters
Abstract
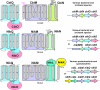
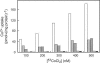
The transition metals nickel and cobalt, essential components of many enzymes, are taken up by specific transport systems of several different types. We integrated in silico and in vivo methods for the analysis of various protein families containing both nickel and cobalt transport systems in prokaryotes. For functional annotation of genes, we used two comparative genomic approaches: identification of regulatory signals and analysis of the genomic positions of genes encoding candidate nickel/cobalt transporters. The nickel-responsive repressor NikR regulates many nickel uptake systems, though the NikR-binding signal is divergent in various taxonomic groups of bacteria and archaea. B(12) riboswitches regulate most of the candidate cobalt transporters in bacteria. The nickel/cobalt transporter genes are often colocalized with genes for nickel-dependent or coenzyme B(12) biosynthesis enzymes. Nickel/cobalt transporters of different families, including the previously known NiCoT, UreH, and HupE/UreJ families of secondary systems and the NikABCDE ABC-type transporters, showed a mosaic distribution in prokaryotic genomes. In silico analyses identified CbiMNQO and NikMNQO as the most widespread groups of microbial transporters for cobalt and nickel ions. These unusual uptake systems contain an ABC protein (CbiO or NikO) but lack an extracytoplasmic solute-binding protein. Experimental analysis confirmed metal transport activity for three members of this family and demonstrated significant activity for a basic module (CbiMN) of the Salmonella enterica serovar Typhimurium transporter.
Figures




References
-
- Abouhamad, W. N., M. Manson, M. M. Gibson, and C. F. Higgins. 1991. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol. Microbiol. 5:1035-1047. - PubMed
-
- Altschul, S. F., and E. V. Koonin. 1998. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 23:444-447. - PubMed
-
- Baginsky, C., J. M. Palacios, J. Imperial, T. Ruiz-Argueso, and B. Brito. 2004. Molecular and functional characterization of the Azorhizobium caulinodans ORS571 hydrogenase gene cluster. FEMS Microbiol. Lett. 237:399-405. - PubMed
-
- Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

