Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences
- PMID: 16354678
- PMCID: PMC1317637
- DOI: 10.1128/MCB.26.1.39-49.2006
Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences
Abstract
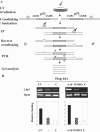
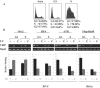
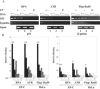
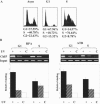
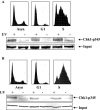
We developed a chromatin immunoprecipitation method for analyzing the binding of repair and checkpoint proteins to DNA base lesions in any region of the human genome. Using this method, we investigated the recruitment of DNA damage checkpoint proteins RPA, Rad9, and ATR to base damage induced by UV and acetoxyacetylaminofluorene in transcribed and nontranscribed regions in wild-type and excision repair-deficient human cells in G1 and S phases of the cell cycle. We find that all 3 damage sensors tested assemble at the site or in the vicinity of damage in the absence of DNA replication or repair and that transcription enhances recruitment of checkpoint proteins to the damage site. Furthermore, we find that UV irradiation of human cells defective in excision repair leads to phosphorylation of Chk1 kinase in both G1 and S phase of the cell cycle, suggesting that primary DNA lesions as well as stalled transcription complexes may act as signals to initiate the DNA damage checkpoint response.
Figures


Similar articles
-
UV-induced G2 checkpoint depends on p38 MAPK and minimal activation of ATR-Chk1 pathway.J Cell Sci. 2013 May 1;126(Pt 9):1923-30. doi: 10.1242/jcs.118265. Epub 2013 Feb 27. J Cell Sci. 2013. PMID: 23447674
-
Protein phosphatase 5 is required for ATR-mediated checkpoint activation.Mol Cell Biol. 2005 Nov;25(22):9910-9. doi: 10.1128/MCB.25.22.9910-9919.2005. Mol Cell Biol. 2005. PMID: 16260606 Free PMC article.
-
Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling.EMBO J. 2006 Jun 7;25(11):2605-14. doi: 10.1038/sj.emboj.7601123. Epub 2006 May 4. EMBO J. 2006. PMID: 16675950 Free PMC article.
-
Chk1 in the DNA damage response: conserved roles from yeasts to mammals.DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1025-32. doi: 10.1016/j.dnarep.2004.03.003. DNA Repair (Amst). 2004. PMID: 15279789 Review.
-
DNA damage checkpoint, damage repair, and genome stability.Yi Chuan Xue Bao. 2006 May;33(5):381-90. doi: 10.1016/S0379-4172(06)60064-4. Yi Chuan Xue Bao. 2006. PMID: 16722332 Review.
Cited by
-
AKAP12 mediates PKA-induced phosphorylation of ATR to enhance nucleotide excision repair.Nucleic Acids Res. 2016 Dec 15;44(22):10711-10726. doi: 10.1093/nar/gkw871. Epub 2016 Sep 28. Nucleic Acids Res. 2016. Retraction in: Nucleic Acids Res. 2020 Nov 18;48(20):11814. doi: 10.1093/nar/gkaa984. PMID: 27683220 Free PMC article. Retracted.
-
Acetaldehyde makes a distinct mutation signature in single-stranded DNA.Nucleic Acids Res. 2022 Jul 22;50(13):7451-7464. doi: 10.1093/nar/gkac570. Nucleic Acids Res. 2022. PMID: 35776120 Free PMC article.
-
ATR kinase inhibition sensitizes quiescent human cells to the lethal effects of cisplatin but increases mutagenesis.Mutat Res. 2019 Nov;816-818:111678. doi: 10.1016/j.mrfmmm.2019.111678. Epub 2019 Sep 17. Mutat Res. 2019. PMID: 31557599 Free PMC article.
-
Ligand modulation of a dinuclear platinum compound leads to mechanistic differences in cell cycle progression and arrest.Biochem Pharmacol. 2013 Dec 15;86(12):1708-20. doi: 10.1016/j.bcp.2013.10.012. Epub 2013 Oct 24. Biochem Pharmacol. 2013. PMID: 24161784 Free PMC article.
-
Reconstitution of a human ATR-mediated checkpoint response to damaged DNA.Proc Natl Acad Sci U S A. 2007 Aug 14;104(33):13301-6. doi: 10.1073/pnas.0706013104. Epub 2007 Aug 8. Proc Natl Acad Sci U S A. 2007. PMID: 17686975 Free PMC article.
References
-
- Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. - PubMed
-
- Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimmer dissociation. Nature 421:499-506. - PubMed
-
- Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. - PubMed
-
- Bermudez, V. P., L. A. Lindsey-Boltz, A. J. Cesare, Y. Maniwa, J. D. Griffith, J. Hurwitz, and A. Sancar. 2003. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA 100:1633-1638. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
