Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4
- PMID: 16354690
- PMCID: PMC1317622
- DOI: 10.1128/MCB.26.1.192-198.2006
Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4
Abstract
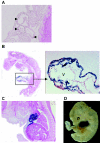
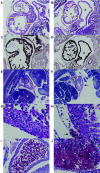
The function of the p53 tumor suppressor to inhibit proliferation or initiate apoptosis is often abrogated in tumor cells. Mdm2 and its homolog, Mdm4, are critical inhibitors of p53 that are often overexpressed in human tumors. In mice, loss of Mdm2 or Mdm4 leads to embryonic lethal phenotypes that are completely rescued by concomitant loss of p53. To examine the role of Mdm2 and Mdm4 in a temporal and tissue-specific manner and to determine the relationships of these inhibitors to each other, we generated conditional alleles. We deleted Mdm2 and Mdm4 in cardiomyocytes, since proliferation and apoptosis are important processes in heart development. Mice lacking Mdm2 in the heart were embryonic lethal and showed defects at the time recombination occurred. A critical number of cardiomyocytes were lost by embryonic day 13.5, resulting in heart failure. This phenotype was completely rescued by deletion of p53. Mice lacking Mdm4 in the heart were born at the correct ratio and appeared to be normal. Our studies provide the first direct evidence that Mdm2 can function in the absence of Mdm4 to regulate p53 activity in a tissue-specific manner. Moreover, Mdm4 cannot compensate for the loss of Mdm2 in heart development.
Figures





References
-
- Agah, R., P. A. Frenkel, B. A. French, L. H. Michael, P. A. Overbeek, and M. D. Schneider. 1997. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Investig. 100:169-179. - PMC - PubMed
-
- Bottger, V., A. Bottger, C. Garcia-Echeverria, Y. F. Ramos, A. J. van der Eb, A. G. Jochemsen, and D. P. Lane. 1999. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene 18:189-199. - PubMed
-
- Chavez-Reyes, A., J. M. Parant, L. L. Amelse, R. M. de Oca Luna, S. J. Korsmeyer, and G. Lozano. 2003. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res. 63:8664-8669. - PubMed
-
- Farley, F. W., P. Soriano, L. S. Steffen, and S. M. Dymecki. 2000. Widespread recombinase expression using FLPeR (Flipper) mice. Genesis 28:106-110. - PubMed
-
- Fernandez, E., Z. Siddiquee, and R. V. Shohet. 2001. Apoptosis and proliferation in the neonatal murine heart. Dev. Dyn. 221:302-310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
