Revealing posttranscriptional regulatory elements through network-level conservation
- PMID: 16355253
- PMCID: PMC1309705
- DOI: 10.1371/journal.pcbi.0010069
Revealing posttranscriptional regulatory elements through network-level conservation
Abstract
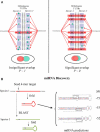
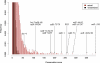
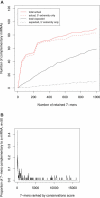
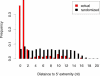
We used network-level conservation between pairs of fly (Drosophila melanogaster/D. pseudoobscura) and worm (Caenorhabditis elegans/C. briggsae) genomes to detect highly conserved mRNA motifs in 3' untranslated regions. Many of these elements are complementary to the 5' extremity of known microRNAs (miRNAs), and likely correspond to their target sites. We also identify known targets of RNA-binding proteins, and many novel sites not yet known to be functional. Coherent sets of genes with similar function often bear the same conserved elements, providing new insights into their cellular functions. We also show that target sites for distinct miRNAs are often simultaneously conserved, suggesting combinatorial regulation by multiple miRNAs. A genome-wide search for conserved stem-loops, containing complementary sequences to the novel sites, revealed many new candidate miRNAs that likely target them. We also provide evidence that posttranscriptional networks have undergone extensive rewiring across distant phyla, despite strong conservation of regulatory elements themselves.
Conflict of interest statement
Figures
References
-
- Lee R, Feinbaum R, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Hutvagner G, Zamore P. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. - PubMed
-
- Rhoades M, Reinhart B, Lim L, Burge C, Bartel B, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. - PubMed
-
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

