Folding free energies of 5'-UTRs impact post-transcriptional regulation on a genomic scale in yeast
- PMID: 16355254
- PMCID: PMC1309706
- DOI: 10.1371/journal.pcbi.0010072
Folding free energies of 5'-UTRs impact post-transcriptional regulation on a genomic scale in yeast
Abstract
Using high-throughput technologies, abundances and other features of genes and proteins have been measured on a genome-wide scale in Saccharomyces cerevisiae. In contrast, secondary structure in 5'-untranslated regions (UTRs) of mRNA has only been investigated for a limited number of genes. Here, the aim is to study genome-wide regulatory effects of mRNA 5'-UTR folding free energies. We performed computations of secondary structures in 5'-UTRs and their folding free energies for all verified genes in S. cerevisiae. We found significant correlations between folding free energies of 5'-UTRs and various transcript features measured in genome-wide studies of yeast. In particular, mRNAs with weakly folded 5'-UTRs have higher translation rates, higher abundances of the corresponding proteins, longer half-lives, and higher numbers of transcripts, and are upregulated after heat shock. Furthermore, 5'-UTRs have significantly higher folding free energies than other genomic regions and randomized sequences. We also found a positive correlation between transcript half-life and ribosome occupancy that is more pronounced for short-lived transcripts, which supports a picture of competition between translation and degradation. Among the genes with strongly folded 5'-UTRs, there is a huge overrepresentation of uncharacterized open reading frames. Based on our analysis, we conclude that (i) there is a widespread bias for 5'-UTRs to be weakly folded, (ii) folding free energies of 5'-UTRs are correlated with mRNA translation and turnover on a genomic scale, and (iii) transcripts with strongly folded 5'-UTRs are often rare and hard to find experimentally.
Conflict of interest statement
Figures
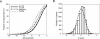
Similar articles
-
Post-termination ribosome interactions with the 5'UTR modulate yeast mRNA stability.EMBO J. 1999 Jun 1;18(11):3139-52. doi: 10.1093/emboj/18.11.3139. EMBO J. 1999. PMID: 10357825 Free PMC article.
-
Identification of novel RARbeta2 transcript variants with short 5'-UTRs in normal and cancerous breast epithelial cells.Oncogene. 2005 Feb 10;24(7):1296-301. doi: 10.1038/sj.onc.1208284. Oncogene. 2005. PMID: 15558014
-
Poly(A)-tail-promoted translation in yeast: implications for translational control.RNA. 1998 Nov;4(11):1321-31. doi: 10.1017/s1355838298980669. RNA. 1998. PMID: 9814754 Free PMC article.
-
Searching for IRES.RNA. 2006 Oct;12(10):1755-85. doi: 10.1261/rna.157806. Epub 2006 Sep 6. RNA. 2006. PMID: 16957278 Free PMC article. Review.
-
Regulation of fungal gene expression via short open reading frames in the mRNA 5'untranslated region.Mol Microbiol. 2003 Aug;49(4):859-67. doi: 10.1046/j.1365-2958.2003.03622.x. Mol Microbiol. 2003. PMID: 12890013 Review.
Cited by
-
Analysis of the Compositional Features and Codon Usage Pattern of Genes Involved in Human Autophagy.Cells. 2022 Oct 12;11(20):3203. doi: 10.3390/cells11203203. Cells. 2022. PMID: 36291071 Free PMC article.
-
Tailor made: the art of therapeutic mRNA design.Nat Rev Drug Discov. 2024 Jan;23(1):67-83. doi: 10.1038/s41573-023-00827-x. Epub 2023 Nov 29. Nat Rev Drug Discov. 2024. PMID: 38030688 Review.
-
The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr 1;3(2):1-35. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 25893143 Free PMC article.
-
The regulatory landscape of 5' UTRs in translational control during zebrafish embryogenesis.bioRxiv [Preprint]. 2023 Nov 23:2023.11.23.568470. doi: 10.1101/2023.11.23.568470. bioRxiv. 2023. Update in: Dev Cell. 2025 May 19;60(10):1498-1515.e8. doi: 10.1016/j.devcel.2024.12.038. PMID: 38045294 Free PMC article. Updated. Preprint.
-
Ribo-Seq Analysis-Based Elucidation of the Dynamic Translation Landscape of Yak Ovarian Tissues in Different Reproductive Stages.FASEB J. 2025 Aug 15;39(15):e70924. doi: 10.1096/fj.202500646RR. FASEB J. 2025. PMID: 40779359 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

