Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm
- PMID: 16356265
- PMCID: PMC1414076
- DOI: 10.1186/gb-2005-6-12-r102
Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm
Erratum in
-
Correction to: Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm.Genome Biol. 2019 Aug 9;20(1):160. doi: 10.1186/s13059-019-1779-z. Genome Biol. 2019. PMID: 31399122 Free PMC article.
Abstract
Background: The male-specific region of the mouse Y chromosome long arm (MSYq) is comprised largely of repeated DNA, including multiple copies of the spermatid-expressed Ssty gene family. Large deletions of MSYq are associated with sperm head defects for which Ssty deficiency has been presumed to be responsible.
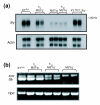
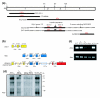
Results: In a search for further candidate genes associated with these defects we analyzed changes in the testis transcriptome resulting from MSYq deletions, using testis cDNA microarrays. This approach, aided by accumulating mouse MSYq sequence information, identified transcripts derived from two further spermatid-expressed multicopy MSYq gene families; like Ssty, each of these new MSYq gene families has multicopy relatives on the X chromosome. The Sly family encodes a protein with homology to the chromatin-associated proteins XLR and XMR that are encoded by the X chromosomal relatives. The second MSYq gene family was identified because the transcripts hybridized to a microarrayed X chromosome-encoded testis cDNA. The X loci ('Astx') encoding this cDNA had 92-94% sequence identity to over 100 putative Y loci ('Asty') across exons and introns; only low level Asty transcription was detected. More strongly transcribed recombinant loci were identified that included Asty exons 2-4 preceded by Ssty1 exons 1, 2 and part of exon 3. Transcription from the Ssty1 promotor generated spermatid-specific transcripts that, in addition to the variable inclusion of Ssty1 and Asty exons, included additional exons because of the serendipitous presence of splice sites further downstream.
Conclusion: We identified further MSYq-encoded transcripts expressed in spermatids and deriving from multicopy Y genes, deficiency of which may underlie the defects in sperm development associated with MSYq deletions.
Figures
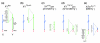






References
-
- Ma K, Inglis JD, Sharkey A, Bickmore WA, Hill RE, Prosser EJ, Speed RM, Thomson EJ, Jobling M, Taylor K, et al. A Y chromosome gene family with RNA-binding protein homology:candidates for the Azoospermia factor AZF controlling human spermatogenesis. Cell. 1993;75:1287–1295. doi: 10.1016/0092-8674(93)90616-X. - DOI - PubMed
-
- Mahadevaiah SK, Odorisio T, Elliott DJ, Rattigan A, Szot M, Laval SH, Washburn LL, McCarrey JR, Cattanach BM, Lovell-Badge R, et al. Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y deletion interval associated with a high incidence of sperm abnormalities. Hum Mol Genet. 1998;7:715–727. doi: 10.1093/hmg/7.4.715. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

