The design of transcription-factor binding sites is affected by combinatorial regulation
- PMID: 16356266
- PMCID: PMC1414079
- DOI: 10.1186/gb-2005-6-12-r103
The design of transcription-factor binding sites is affected by combinatorial regulation
Abstract
Background: Transcription factors regulate gene expression by binding to specific cis-regulatory elements in gene promoters. Although DNA sequences that serve as transcription-factor binding sites have been characterized and associated with the regulation of numerous genes, the principles that govern the design and evolution of such sites are poorly understood.
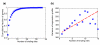
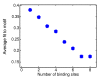
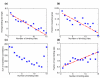
Results: Using the comprehensive mapping of binding-site locations available in Saccharomyces cerevisiae, we examined possible factors that may have an impact on binding-site design. We found that binding sites tend to be shorter and fuzzier when they appear in promoter regions that bind multiple transcription factors. We further found that essential genes bind relatively fewer transcription factors, as do divergent promoters. We provide evidence that novel binding sites tend to appear in specific promoters that are already associated with multiple sites.
Conclusion: Two principal models may account for the observed correlations. First, it may be that the interaction between multiple factors compensates for the decreased specificity of each specific binding sequence. In such a scenario, binding-site fuzziness is a consequence of the presence of multiple binding sites. Second, binding sites may tend to appear in promoter regions that are subject to low selective pressure, which also allows for fuzzier motifs. The latter possibility may account for the relatively low number of binding sites found in promoters of essential genes and in divergent promoters.
Figures

References
-
- Carey M, Smale ST. Transcriptional Regulation in Eukaryotes. Cold Spring Harbor, New York: CSHL Press; 1999.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

