MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling
- PMID: 16357195
- PMCID: PMC1315275
- DOI: 10.1073/pnas.0509535102
MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling
Abstract
MicroRNAs (miRNAs) are genomically encoded small RNAs that hybridize with messenger RNAs, resulting in degradation or translational inhibition of targeted transcripts. The potential for miRNAs to regulate cell-lineage determination or differentiation from pluripotent progenitor or stem cells is unknown. Here, we show that microRNA1 (miR-1) is an ancient muscle-specific gene conserved in sequence and expression in Drosophila. Drosophila miR-1 (dmiR-1) is regulated through a serum response factor-like binding site in cardiac progenitor cells. Loss- and gain-of-function studies demonstrated a role for dmiR-1 in modulating cardiogenesis and in maintenance of muscle-gene expression. We provide in vivo evidence that dmiR-1 targets transcripts encoding the Notch ligand Delta, providing a potential mechanism for the expansion of cardiac and muscle progenitor cells and failure of progenitor cell differentiation in some dmiR-1 mutants. These findings demonstrate that dmiR-1 may "fine-tune" critical steps involved in differentiation of cardiac and somatic muscle progenitors and targets a pathway required for progenitor cell specification and asymmetric cell division.
Figures
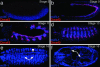
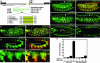
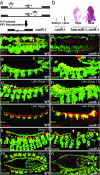
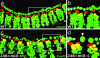

References
-
- Ambros, V. (2001) Cell 107, 823–826. - PubMed
-
- Johnston, R. J. & Hobert, O. (2003) Nature 426, 845–849. - PubMed
-
- Bartel, D. P. (2004) Cell 116, 281–297. - PubMed
-
- Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P. & Burge, C. B. (2003) Cell 115, 787–798. - PubMed
-
- Zhao, Y., Samal, E. & Srivastava, D. (2005) Nature 436, 214–220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

