The global transcriptional regulatory network for metabolism in Escherichia coli exhibits few dominant functional states
- PMID: 16357206
- PMCID: PMC1323155
- DOI: 10.1073/pnas.0505231102
The global transcriptional regulatory network for metabolism in Escherichia coli exhibits few dominant functional states
Abstract
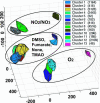
A principal aim of systems biology is to develop in silico models of whole cells or cellular processes that explain and predict observable cellular phenotypes. Here, we use a model of a genome-scale reconstruction of the integrated metabolic and transcriptional regulatory networks for Escherichia coli, composed of 1,010 gene products, to assess the properties of all functional states computed in 15,580 different growth environments. The set of all functional states of the integrated network exhibits a discernable structure that can be visualized in 3-dimensional space, showing that the transcriptional regulatory network governing metabolism in E. coli responds primarily to the available electron acceptor and the presence of glucose as the carbon source. This result is consistent with recently published experimental data. The observation that a complex network composed of 1,010 genes is organized to achieve few dominant modes demonstrates the utility of the systems approach for consolidating large amounts of genome-scale molecular information about a genome and its regulation to elucidate an organism's preferred environments and functional capabilities.
Figures


References
-
- Selinger, D. W., Wright, M. A. & Church, G. M. (2003) Trends Biotechnol. 21, 251-254. - PubMed
-
- Ideker, T., Galitski, T. & Hood, L. (2001) Annu. Rev. Genom. Hum. Genet. 2, 343-372. - PubMed
-
- Kitano, H. (2002) Science 295, 1662-1664. - PubMed
-
- Kitano, H. (2002) Nature 420, 206-210. - PubMed
-
- Oltvai, Z. N. & Barabasi, A. L. (2002) Science 298, 763-764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

