Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans
- PMID: 16360034
- PMCID: PMC4435800
- DOI: 10.1016/j.cell.2005.09.034
Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans
Abstract
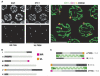
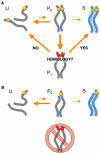
We have investigated the role of pairing centers (PCs), cis-acting sites required for accurate segregation of homologous chromosomes during meiosis in C. elegans. We find that these sites play two distinct roles that contribute to proper segregation. Chromosomes lacking PCs usually fail to synapse and also lack a synapsis-independent stabilization activity. The presence of a PC on just one copy of a chromosome pair promotes synapsis but does not support synapsis-independent pairing stabilization, indicating that these functions are separable. Once initiated, synapsis is highly processive, even between nonhomologous chromosomes of disparate lengths, elucidating how translocations suppress meiotic recombination in C. elegans. These findings suggest a multistep pathway for chromosome synapsis in which PCs impart selectivity and efficiency through a "kinetic proofreading" mechanism. We speculate that concentration of these activities at one region per chromosome may have coevolved with the loss of a point centromere to safeguard karyotype stability.
Figures
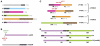
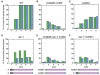

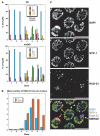
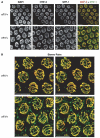
Comment in
-
When size does not matter: pairing sites during meiosis.Cell. 2005 Dec 16;123(6):989-92. doi: 10.1016/j.cell.2005.11.031. Cell. 2005. PMID: 16360029 Review. No abstract available.
References
-
- Albertson DG, Rose AM, Villeneuve AM. Chromosome organization, mitosis, and meiosis. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. pp. 47–78. - PubMed
-
- Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–474. - PubMed
-
- Couteau F, Nabeshima K, Villeneuve A, Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 2004;14:585–592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

