Fold recognition of the human immunodeficiency virus type 1 V3 loop and flexibility of its crown structure during the course of adaptation to a host
- PMID: 16361230
- PMCID: PMC1456290
- DOI: 10.1534/genetics.105.051508
Fold recognition of the human immunodeficiency virus type 1 V3 loop and flexibility of its crown structure during the course of adaptation to a host
Abstract
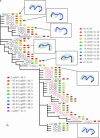
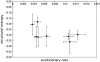
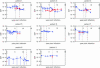
The third hypervariable (V3) region of the HIV-1 gp120 protein is responsible for many aspects of viral infectivity. The tertiary structure of the V3 loop seems to influence the coreceptor usage of the virus, which is an important determinant of HIV pathogenesis. Hence, the information about preferred conformations of the V3-loop region and its flexibility could be a crucial tool for understanding the mechanisms of progression from an initial infection to AIDS. Taking into account the uncertainty of the loop structure, we predicted the structural flexibility, diversity, and sequence fitness to the V3-loop structure for each of the sequences serially sampled during an asymptomatic period. Structural diversity correlated with sequence diversity. The predicted crown structure usage implied that structural flexibility depended on the patient and that the antigenic character of the virus might be almost uniform in a patient whose immune system is strong. Furthermore, the predicted structural ensemble suggested that toward the end of the asymptomatic period there was a change in the V3-loop structure or in the environment surrounding the V3 loop, possibly because of its proximity to the gp120 core.
Figures
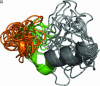
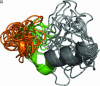
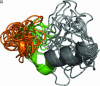




References
-
- Baker, D., 2000. A surprising simplicity to protein folding. Nature 405: 39–42. - PubMed
-
- Baker, D., and A. Sali, 2001. Protein structure prediction and structural genomics. Science 294: 93–96. - PubMed
-
- Balbach, J. J., J. Yang, D. P. Weliky, P. J. Steinbach, V. Tugarinov et al., 2000. Probing hydrogen bonds in the antibody-bound HIV-1 gp120 V3 loop by solid state NMR REDOR measurements. J. Biomol. NMR 16: 313–327. - PubMed
-
- Berger, E. A., R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman et al., 1998. A new classification for HIV-1. Nature 391: 240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

