Human and mouse oligonucleotide-based array CGH
- PMID: 16361265
- PMCID: PMC1316119
- DOI: 10.1093/nar/gni191
Human and mouse oligonucleotide-based array CGH
Abstract
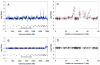
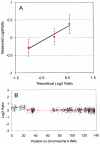
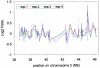
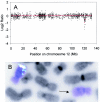
Array-based comparative genomic hybridization is a high resolution method for measuring chromosomal copy number changes. Here we present a validated protocol using in-house spotted oligonucleotide libraries for array comparative genomic hybridization (CGH). This oligo array CGH platform yields reproducible results and is capable of detecting single copy gains, multi-copy amplifications as well as homozygous and heterozygous deletions as small as 100 kb with high resolution. A human oligonucleotide library was printed on amine binding slides. Arrays were hybridized using a hybstation and analysed using BlueFuse feature extraction software, with >95% of spots passing quality control. The protocol allows as little as 300 ng of input DNA and a 90% reduction of Cot-1 DNA without compromising quality. High quality results have also been obtained with DNA from archival tissue. Finally, in addition to human oligo arrays, we have applied the protocol successfully to mouse oligo arrays. We believe that this oligo-based platform using 'off-the-shelf' oligo libraries provides an easy accessible alternative to BAC arrays for CGH, which is cost-effective, available at high resolution and easily implemented for any sequenced organism without compromising the quality of the results.
Figures

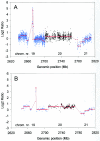
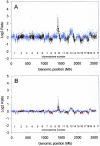
References
-
- Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genet. 1998;20:207–211. - PubMed
-
- Oostlander A.E., Meijer G.A., Ylstra B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin. Genet. 2004;66:488–495. - PubMed
-
- Snijders A.M., Nowak N., Segraves R., Blackwood S., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nature Genet. 2001;29:263–264. - PubMed
-
- Kallioniemi A., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. - PubMed
-
- Ishkanian A.S., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nature Genet. 2004;36:299–303. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

