DNA base flipping by a base pair-mimic nucleoside
- PMID: 16361269
- PMCID: PMC1316115
- DOI: 10.1093/nar/gki1018
DNA base flipping by a base pair-mimic nucleoside
Abstract
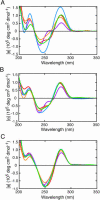
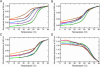
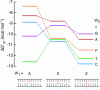
On the basis of non-covalent bond interactions in nucleic acids, we synthesized the deoxyadenosine derivatives tethering a phenyl group (X) and a naphthyl group (Z) by an amide linker, which mimic a Watson-Crick base pair. Circular dichroism spectra indicated that the duplexes containing X and Z formed a similar conformation regardless of the opposite nucleotide species (A, G, C, T and an abasic site analogue F), which was not observed for the natural duplexes. The values among the natural duplexes containing the A/A, A/G, A/C, A/T and A/F pairs differed by 5.2 kcal mol(-1) while that among the duplexes containing X or Z in place of the adenine differed by only 1.9 or 2.8 kcal mol(-1), respectively. Fluorescence quenching experiments confirmed that 2-amino purine opposite X adopted an unstacked conformation. The structural and thermodynamic analyses suggest that the aromatic hydrocarbon group of X and Z intercalates into a double helix, resulting in the opposite nucleotide base flipping into an unstacked position regardless of the nucleotide species. This observation implies that modifications at the aromatic hydrocarbon group and the amide linker may expand the application of the base pair-mimic nucleosides for molecular biology and biotechnology.
Figures
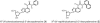

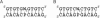
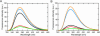
Similar articles
-
Dynamics and energetics of the base flipping conformation studied with base pair-mimic nucleosides.Biochemistry. 2009 Dec 1;48(47):11304-11. doi: 10.1021/bi901496q. Biochemistry. 2009. PMID: 19839646
-
Functionalization of DNA by the base pair-mimic nucleosides.Nucleic Acids Symp Ser (Oxf). 2007;(51):151-2. doi: 10.1093/nass/nrm076. Nucleic Acids Symp Ser (Oxf). 2007. PMID: 18029631
-
Site-selective RNA cleavage by DNA bearing a base pair-mimic nucleoside.J Am Chem Soc. 2005 Jan 19;127(2):518-9. doi: 10.1021/ja045445s. J Am Chem Soc. 2005. PMID: 15643864
-
Expanded-size bases in naturally sized DNA: evaluation of steric effects in Watson-Crick pairing.J Am Chem Soc. 2004 Sep 29;126(38):11826-31. doi: 10.1021/ja048499a. J Am Chem Soc. 2004. PMID: 15382917
-
Large stacking stability of a base pair-mimic nucleotide on the DNA duplex.Nucleic Acids Res Suppl. 2003;(3):79-80. doi: 10.1093/nass/3.1.79. Nucleic Acids Res Suppl. 2003. PMID: 14510389
Cited by
-
Conformational changes of the phenyl and naphthyl isocyanate-DNA adducts during DNA replication and by minor groove binding molecules.Nucleic Acids Res. 2013 Oct;41(18):8581-90. doi: 10.1093/nar/gkt608. Epub 2013 Jul 19. Nucleic Acids Res. 2013. PMID: 23873956 Free PMC article.
-
Selective alkylation of T-T mismatched DNA using vinyldiaminotriazine-acridine conjugate.Nucleic Acids Res. 2018 Feb 16;46(3):1059-1068. doi: 10.1093/nar/gkx1278. Nucleic Acids Res. 2018. PMID: 29309639 Free PMC article.
-
Use of nucleic Acid analogs for the study of nucleic Acid interactions.J Nucleic Acids. 2011;2011:967098. doi: 10.4061/2011/967098. Epub 2011 Jul 24. J Nucleic Acids. 2011. PMID: 21822475 Free PMC article.
-
A molecular dynamics study of slow base flipping in DNA using conformational flooding.Biophys J. 2007 Aug 1;93(3):770-86. doi: 10.1529/biophysj.106.091751. Epub 2007 May 11. Biophys J. 2007. PMID: 17496048 Free PMC article.
-
Fluorescent analogs of biomolecular building blocks: design, properties, and applications.Chem Rev. 2010 May 12;110(5):2579-619. doi: 10.1021/cr900301e. Chem Rev. 2010. PMID: 20205430 Free PMC article. Review. No abstract available.
References
-
- Turner D.H., Sugimoto N., Kierzek R., Dreiker S.D. Free energy increments for hydrogen bonds in nucleic acid base pairs. J. Am. Chem. Soc. 1987;109:3783–3784.
-
- Moody E.M., Bevilacqua P.C. Folding of a stable DNA motif involves a highly cooperative network of interactions. J. Am. Chem. Soc. 2003;125:16285–16293. - PubMed
-
- Morales J.C., Kool E.T. Functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. - PubMed
-
- Kool E.T., Morales J.C., Guckian K.M. Mimicking the structure and function of DNA: insights into DNA stability and replication. Angew. Chem. Int. Ed. Engl. 2000;39:990–1099. - PubMed

