Automatic assessment of alignment quality
- PMID: 16361270
- PMCID: PMC1316116
- DOI: 10.1093/nar/gki1020
Automatic assessment of alignment quality
Abstract
Multiple sequence alignments play a central role in the annotation of novel genomes. Given the biological and computational complexity of this task, the automatic generation of high-quality alignments remains challenging. Since multiple alignments are usually employed at the very start of data analysis pipelines, it is crucial to ensure high alignment quality. We describe a simple, yet elegant, solution to assess the biological accuracy of alignments automatically. Our approach is based on the comparison of several alignments of the same sequences. We introduce two functions to compare alignments: the average overlap score and the multiple overlap score. The former identifies difficult alignment cases by expressing the similarity among several alignments, while the latter estimates the biological correctness of individual alignments. We implemented both functions in the MUMSA program and demonstrate the overall robustness and accuracy of both functions on three large benchmark sets.
Figures
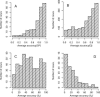

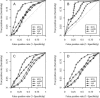
References
-
- Lecompte O., Thompson J.D., Plewniak F., Thierry J., Poch O. Multiple alignment of complete sequences (MACS) in the postgenomic era. Gene. 2001;270:17–30. - PubMed
-
- Van Walle I., Lasters I., Wyns L. Align-m—a new algorithm for multiple alignment of highly divergent sequences. Bioinformatics. 2004;20:1428–1435. - PubMed

