The Arabidopsis group 1 LATE EMBRYOGENESIS ABUNDANT protein ATEM6 is required for normal seed development
- PMID: 16361514
- PMCID: PMC1326038
- DOI: 10.1104/pp.105.072967
The Arabidopsis group 1 LATE EMBRYOGENESIS ABUNDANT protein ATEM6 is required for normal seed development
Abstract
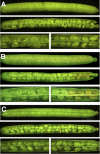
As part of the embryo maturation process, orthodox seeds undergo a developmentally regulated dehydration period. The LATE EMBRYOGENESIS ABUNDANT (LEA) genes encode a large and diverse family of proteins expressed during this time. Many hypothesize that LEA proteins act by mitigating water loss and maintaining cellular stability within the desiccated seed, although the mechanisms of their actions remain largely unknown. The model plant Arabidopsis (Arabidopsis thaliana) contains two genes belonging to the group 1 LEA family, ATEM1 and ATEM6, and knockout mutations in these genes are being sought as a means to better understand group 1 LEA protein function during embryo maturation. We have identified a T-DNA insertion allele of the ATEM6 gene in which the T-DNA is present just downstream of the protein coding region. While this gene is transcriptionally active and encodes a wild-type protein, there is no detectable ATEM6 protein in mature seeds. Mutant seeds display premature seed dehydration and maturation at the distal end of siliques, demonstrating that this protein is required for normal seed development. We propose that one function for group 1 LEA proteins in seed development is to buffer the water loss that occurs during embryo maturation and that loss of ATEM6 expression results in the mutant phenotype.
Figures




Similar articles
-
Seed dehydration and the establishment of desiccation tolerance during seed maturation is altered in the Arabidopsis thaliana mutant atem6-1.Plant Cell Physiol. 2009 Feb;50(2):243-53. doi: 10.1093/pcp/pcn185. Epub 2008 Dec 10. Plant Cell Physiol. 2009. PMID: 19073649
-
LEC1, FUS3, ABI3 and Em expression reveals no correlation with dormancy in Arabidopsis.J Exp Bot. 2004 Jan;55(394):77-87. doi: 10.1093/jxb/erh014. J Exp Bot. 2004. PMID: 14676287
-
Differential expression of the Arabidopsis genes coding for Em-like proteins.J Exp Bot. 2000 Jul;51(348):1211-20. J Exp Bot. 2000. PMID: 10937696
-
Genetic activity during early plant embryogenesis.Biochem J. 2020 Oct 16;477(19):3743-3767. doi: 10.1042/BCJ20190161. Biochem J. 2020. PMID: 33045058 Free PMC article. Review.
-
Seed development: with or without sex?Curr Biol. 1999 Sep 9;9(17):R636-9. doi: 10.1016/s0960-9822(99)80411-4. Curr Biol. 1999. PMID: 10508576 Review.
Cited by
-
TMT-Based Quantitative Proteomic Analysis Reveals the Physiological Regulatory Networks of Embryo Dehydration Protection in Lotus (Nelumbo nucifera).Front Plant Sci. 2021 Dec 17;12:792057. doi: 10.3389/fpls.2021.792057. eCollection 2021. Front Plant Sci. 2021. PMID: 34975978 Free PMC article.
-
ABI5-FLZ13 module transcriptionally represses growth-related genes to delay seed germination in response to ABA.Plant Commun. 2023 Nov 13;4(6):100636. doi: 10.1016/j.xplc.2023.100636. Epub 2023 Jun 9. Plant Commun. 2023. PMID: 37301981 Free PMC article.
-
Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):3979-84. doi: 10.1073/pnas.1301009110. Epub 2013 Feb 19. Proc Natl Acad Sci U S A. 2013. PMID: 23431200 Free PMC article.
-
Identification and characterization of long non-coding RNA (lncRNA) in the developing seeds of Jatropha curcas.Sci Rep. 2020 Jun 25;10(1):10395. doi: 10.1038/s41598-020-67410-x. Sci Rep. 2020. PMID: 32587349 Free PMC article.
-
Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum).Planta. 2015 Mar;241(3):757-72. doi: 10.1007/s00425-014-2215-y. Epub 2014 Dec 10. Planta. 2015. PMID: 25491641
References
-
- Baker J, Steele C, Dure L III (1988) Sequence and characterization of 6 Lea proteins and their genes from cotton. Plant Mol Biol 11: 277–291 - PubMed
-
- Bartels D, Singh M, Salamini F (1988) Onset of desiccation tolerance during development of the barley embryo. Planta 175: 485–492 - PubMed
-
- Berge SK, Bartholomew DM, Quatrano RS (1989) Control of the expression of wheat embryo genes by abscisic acid. In RL Goldberg, ed, Molecular Basis of Plant Development. Alan R. Liss, New York, pp 193–201
-
- Bies N, Aspart L, Carles C, Gallois P, Delseny M (1998) Accumulation and degradation of Em proteins in Arabidopsis thaliana: evidence for post-transcriptional controls. J Exp Bot 49: 1925–1933
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

