A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation
- PMID: 16362040
- PMCID: PMC1356358
- DOI: 10.1038/sj.emboj.7600915
A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation
Abstract
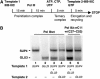
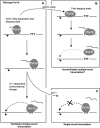
While initiation of transcription by RNA polymerase III (Pol III) has been thoroughly investigated, molecular mechanisms driving transcription termination remain poorly understood. Here we describe how the characterization of the in vitro transcriptional properties of a Pol III variant (Pol IIIdelta), lacking the C11, C37, and C53 subunits, revealed crucial information about the mechanisms of Pol III termination and reinitiation. The specific requirement for the C37-C53 complex in terminator recognition was determined. This complex was demonstrated to slow down elongation by the enzyme, adding to the evidence implicating the elongation rate as a critical determinant of correct terminator recognition. In addition, the presence of the C37-C53 complex required the simultaneous addition of C11 to Pol IIIdelta for the enzyme to reinitiate after the first round of transcription, thus uncovering a role for polymerase subunits in the facilitated recycling process. Interestingly, we demonstrated that the role of C11 in recycling was independent of its role in RNA cleavage. The data presented allowed us to propose a model of Pol III termination and its links to reinitiation.
Figures

Similar articles
-
The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening.J Biol Chem. 2010 Jan 22;285(4):2695-706. doi: 10.1074/jbc.M109.074013. Epub 2009 Nov 24. J Biol Chem. 2010. PMID: 19940126 Free PMC article.
-
Distinguishing core and holoenzyme mechanisms of transcription termination by RNA polymerase III.Mol Cell Biol. 2013 Apr;33(8):1571-81. doi: 10.1128/MCB.01733-12. Epub 2013 Feb 11. Mol Cell Biol. 2013. PMID: 23401852 Free PMC article.
-
Mechanism of RNA polymerase III termination-associated reinitiation-recycling conferred by the essential function of the N terminal-and-linker domain of the C11 subunit.Nat Commun. 2021 Oct 8;12(1):5900. doi: 10.1038/s41467-021-26080-7. Nat Commun. 2021. PMID: 34625550 Free PMC article.
-
Transcription termination by the eukaryotic RNA polymerase III.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):318-30. doi: 10.1016/j.bbagrm.2012.10.006. Epub 2012 Oct 23. Biochim Biophys Acta. 2013. PMID: 23099421 Free PMC article. Review.
-
Yeast RNA polymerase III transcription factors and effectors.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):283-95. doi: 10.1016/j.bbagrm.2012.10.002. Epub 2012 Oct 12. Biochim Biophys Acta. 2013. PMID: 23063749 Review.
Cited by
-
A mutation in POLR3E impairs antiviral immune response and RNA polymerase III.Proc Natl Acad Sci U S A. 2020 Sep 8;117(36):22113-22121. doi: 10.1073/pnas.2009947117. Epub 2020 Aug 25. Proc Natl Acad Sci U S A. 2020. PMID: 32843346 Free PMC article.
-
Ancient origin, functional conservation and fast evolution of DNA-dependent RNA polymerase III.Nucleic Acids Res. 2006 Jul 28;34(13):3615-24. doi: 10.1093/nar/gkl421. Print 2006. Nucleic Acids Res. 2006. PMID: 16877568 Free PMC article.
-
The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening.J Biol Chem. 2010 Jan 22;285(4):2695-706. doi: 10.1074/jbc.M109.074013. Epub 2009 Nov 24. J Biol Chem. 2010. PMID: 19940126 Free PMC article.
-
Molecular structures of unbound and transcribing RNA polymerase III.Nature. 2015 Dec 10;528(7581):231-6. doi: 10.1038/nature16143. Epub 2015 Nov 25. Nature. 2015. PMID: 26605533 Free PMC article.
-
Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation.Transcription. 2014;5(1):e27639. doi: 10.4161/trns.27369. Transcription. 2014. PMID: 25764110 Free PMC article. Review.
References
-
- Alen C, Kent NA, Jones HS, O'Sullivan J, Aranda A, Proudfoot NJ (2002) A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell 10: 1441–1452 - PubMed
-
- Awrey DE, Shimasaki N, Koth C, Weilbaecher R, Olmsted V, Kazanis S, Shan X, Arellano J, Arrowsmith CH, Kane CM, Edwards AM (1998) Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J Biol Chem 273: 22595–22605 - PubMed
-
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E (2005) A ratchet mechanism of transcription elongation and its control. Cell 120: 183–193 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

