Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb
- PMID: 16362050
- PMCID: PMC1356324
- DOI: 10.1038/sj.emboj.7600883
Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb
Abstract
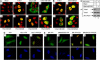
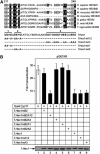
Transcription elongation of eukaryotic genes by RNA polymerase II depends on the positive transcription elongation factor b (P-TEFb). When sequestered into the large complex, P-TEFb kinase activity is inhibited by the coordinate actions of 7SK small nuclear RNA (7SK snRNA) and hexamethylene bisacetamide (HMBA)-induced protein 1 (HEXIM1). We found that the basic region in HEXIM1 directs its nuclear import via two monopartite and two bipartite nuclear localization sequences. Moreover, the arginine-rich motif within it is essential for its binding to 7SK snRNA, P-TEFb, and inhibition of transcription. Notably, the basic region interacts with the adjacent acidic regions in the absence of RNA. The removal of the positive or negative charges from these regions in HEXIM1 leads to its sequestration into the large complex and inhibition of transcription independently of the arginine-rich motif. Finally, the removal of the negative charges from HEXIM1 results in its subnuclear localization into nuclear speckles. We propose a model where the interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct its binding to P-TEFb and subcellular localization that culminates in the inhibition of transcription.
Figures






References
-
- Byers SA, Price JP, Cooper JJ, Li Q, Price DH (2005) HEXIM2, a HEXIM1 related protein, regulates P-TEFb through association with 7SK. J Biol Chem 280: 16360–16367 - PubMed
-
- Carmo-Fonseca M (2002) The contribution of nuclear compartmentalization to gene regulation. Cell 108: 513–521 - PubMed
-
- Chao SH, Price DH (2001) Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276: 31793–31799 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

