Translational control in positive strand RNA plant viruses
- PMID: 16364749
- PMCID: PMC1847782
- DOI: 10.1016/j.virol.2005.09.031
Translational control in positive strand RNA plant viruses
Abstract
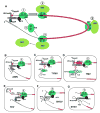
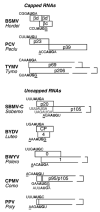
The great variety of genome organizations means that most plant positive strand viral RNAs differ from the standard 5'-cap/3'-poly(A) structure of eukaryotic mRNAs. The cap and poly(A) tail recruit initiation factors that support the formation of a closed loop mRNA conformation, the state in which translation initiation is most efficient. We review the diverse array of cis-acting sequences present in viral mRNAs that compensate for the absence of a cap, poly(A) tail, or both. We also discuss the cis-acting sequences that control translation strategies that both amplify the coding potential of a genome and regulate the accumulations of viral gene products. Such strategies include leaky scanning initiation of translation of overlapping open reading frames, stop codon readthrough, and ribosomal frameshifting. Finally, future directions for research on the translation of plant positive strand viruses are discussed.
Figures

References
-
- Aranda M, Maule A. Virus-induced host gene shutoff in animals and plants. Virology. 1998;243:261–267. - PubMed
-
- Baranov PV, Gesteland RF, Atkins JF. Recording: translational bifurcations in gene expression. Gene. 2002;286:187–201. - PubMed
-
- Barends S, Bink HH, van den Worm SH, Pleij CW, Kraal B. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell. 2003;112:123–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

