Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases
- PMID: 16365295
- PMCID: PMC1316884
- DOI: 10.1073/pnas.0509418102
Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases
Abstract
Ubiquitin-conjugating enzymes (E2s) collaborate with the ubiquitin-activating enzyme (E1) and ubiquitin ligases (E3s) to attach ubiquitin to target proteins. RING-containing E3s simultaneously bind to E2s and substrates, bringing them into close proximity and thus facilitating ubiquitination. We show herein that, although the E3-binding site on the human E2 UbcH5b is distant from its active site, two RING-type minimal E3 modules lacking substrate-binding functions greatly stimulate the rate of ubiquitin release from the UbcH5b-ubiquitin thioester. Using statistical coupling analysis and mutagenesis, we identify and characterize clusters of coevolving and functionally linked residues within UbcH5b that span its E3-binding and active sites. Several UbcH5b mutants are defective in their stimulation by E3s despite their abilities to bind to these E3s, to form ubiquitin thioesters, and to release ubiquitin at a basal rate. One such mutation, I37A, is distant from both the active site and the E3-binding site of UbcH5b. Our studies reveal structural determinants for communication between distal functional sites of E2s and suggest that RING-type E3s activate E2s allosterically.
Figures

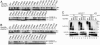

References
-
- Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. - PubMed
-
- Voges, D., Zwickl, P. & Baumeister, W. (1999) Annu. Rev. Biochem. 68, 1015–1068. - PubMed
-
- Pickart, C. M. & Eddins, M. J. (2004) Biochim. Biophys. Acta 1695, 55–72. - PubMed
-
- Bloom, J., Amador, V., Bartolini, F., DeMartino, G. & Pagano, M. (2003) Cell 115, 71–82. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

