Widespread genetic exchange among terrestrial bacteriophages
- PMID: 16365305
- PMCID: PMC1323146
- DOI: 10.1073/pnas.0503074102
Widespread genetic exchange among terrestrial bacteriophages
Abstract
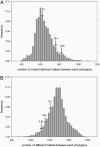
Bacteriophages are the most numerous entities in the biosphere. Despite this numerical dominance, the genetic structure of bacteriophage populations is poorly understood. Here, we present a biogeography study involving 25 previously undescribed bacteriophages from the Cystoviridae clade, a group characterized by a dsRNA genome divided into three segments. Previous laboratory manipulation has shown that, when multiple Cystoviruses infect a single host cell, they undergo (i) rare intrasegment recombination events and (ii) frequent genetic reassortment between segments. Analyzing linkage disequilibrium (LD) within segments, we find no significant evidence of intrasegment recombination in wild populations, consistent with (i). An extensive analysis of LD between segments supports frequent reassortment, on a time scale similar to the genomic mutation rate. The absence of LD within this group of phages is consistent with expectations for a completely sexual population, despite the fact that some segments have >50% nucleotide divergence at 4-fold degenerate sites. This extraordinary rate of genetic exchange between highly unrelated individuals is unprecedented in any taxa. We discuss our results in light of the biological species concept applied to viruses.
Figures

References
-
- Hendrix, R. W. (2002) Theor. Popul. Biol. 61, 471-480. - PubMed
-
- Xu, X. Y., Lindstrom, S. E., Shaw, M. W., Smith, C. B., Hall, H. E., Mungall, B. A., Subbarao, K., Cox, N. J. & Klimov, A. (2004) Virus Res. 103, 55-60. - PubMed
-
- Lindstrom, S. E., Cox, N. J. & Klimov, A. (2004) Virology 328, 101-119. - PubMed
-
- Lin, Y. P., Gregory, V., Bennett, M. & Hay, A. (2004) Virus Res. 103, 47-52. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

