Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host
- PMID: 16365377
- PMCID: PMC1361709
- DOI: 10.1101/gr.4106106
Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host
Abstract
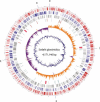
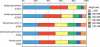
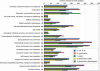
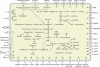
Sodalis glossinidius is a maternally transmitted endosymbiont of tsetse flies (Glossina spp.), an insect of medical and veterinary significance. Analysis of the complete sequence of Sodalis' chromosome (4,171,146 bp, encoding 2,432 protein coding sequences) indicates a reduced coding capacity of 51%. Furthermore, the chromosome contains 972 pseudogenes, an inordinately high number compared with that of other bacterial species. A high proportion of these pseudogenes are homologs of known proteins that function either in defense or in the transport and metabolism of carbohydrates and inorganic ions, suggesting Sodalis' degenerative adaptations to the immunity and restricted nutritional status of the host. Sodalis possesses three chromosomal symbiosis regions (SSR): SSR-1, SSR-2, and SSR-3, with gene inventories similar to the Type-III secretion system (TTSS) ysa from Yersinia enterolitica and SPI-1 and SPI-2 from Salmonella, respectively. While core components of the needle structure have been conserved, some of the effectors and regulators typically associated with these systems in pathogenic microbes are modified or eliminated in Sodalis. Analysis of SSR-specific invA transcript abundance in Sodalis during host development indicates that the individual symbiosis regions may exhibit different temporal expression profiles. In addition, the Sodalis chromosome encodes a complete flagella structure, key components of which are expressed in immature host developmental stages. These features may be important for the transmission and establishment of symbiont infections in the intra-uterine progeny. The data suggest that Sodalis represents an evolutionary intermediate transitioning from a free-living to a mutualistic lifestyle.
Figures

References
-
- Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M., and Aksoy, S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse, Wigglesworthia glossinidia. Nat. Genet. 32 402-407. - PubMed
-
- Aksoy, S. 1995. Molecular analysis of the endosymbionts of tsetse flies: 16S rDNA locus and over-expression of a chaperonin. Insect Mol. Biol. 4 23-29. - PubMed
-
- ———. 2000. Tsetse: A haven for microorganisms. Parasitol. Today 16 114-118. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
