Defining the mammalian CArGome
- PMID: 16365378
- PMCID: PMC1361715
- DOI: 10.1101/gr.4108706
Defining the mammalian CArGome
Abstract
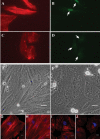
Serum response factor (SRF) binds a 1216-fold degenerate cis element known as the CArG box. CArG boxes are found primarily in muscle- and growth-factor-associated genes although the full spectrum of functional CArG elements in the genome (the CArGome) has yet to be defined. Here we describe a genome-wide screen to further define the functional mammalian CArGome. A computational approach involving comparative genomic analyses of human and mouse orthologous genes uncovered >100 hypothetical SRF-dependent genes, including 10 previously identified SRF targets, harboring a conserved CArG element within 4000 bp of the annotated transcription start site (TSS). We PCR-cloned 89 hypothetical SRF targets and subjected each of them to at least two of several validations including luciferase reporter, gel shift, chromatin immunoprecipitation, and mRNA expression following RNAi knockdown of SRF; 60/89 (67%) of the targets were validated. Interestingly, 26 of the validated SRF target genes encode for cytoskeletal/contractile or adhesion proteins. RNAi knockdown of SRF diminishes expression of several SRF-dependent cytoskeletal genes and elicits an attending perturbation in the cytoarchitecture of both human and rodent cells. These data illustrate the power of integrating existing algorithms to interrogate the genome in a relatively unbiased fashion for cis-regulatory element discovery. In this manner, we have further expanded the mammalian CArGome with the discovery of an array of cyto-contractile genes that coordinate normal cytoskeletal homeostasis. We suggest one function of SRF is that of an ancient master regulator of the actin cytoskeleton.
Figures



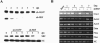


References
-
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299 1391-1394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
