A genome-wide study of dual coding regions in human alternatively spliced genes
- PMID: 16365380
- PMCID: PMC1361714
- DOI: 10.1101/gr.4246506
A genome-wide study of dual coding regions in human alternatively spliced genes
Abstract
Alternative splicing is a major mechanism for gene product regulation in many multicellular organisms. By using different exon combinations, some coding regions can encode amino acids in multiple reading frames in different transcripts. Here we performed a systematic search through a set of high-quality human transcripts and show that approximately 7% of alternatively spliced genes contain dual (multiple) coding regions. By using a conservative criterion, we found that in these regions most secondary reading frames evolved recently in mammals, and a significant proportion of them may be specific to primates. Based on the presence of in-frame stop codons in orthologous sequences in other animals, we further classified ancestral and derived reading frames in these regions. Our results indicated that ancestral reading frames are usually under stronger selection than are derived reading frames. Ancestral reading frames mainly influence the coding properties of these dual coding regions. Compared with coding regions of the whole genome, ancestral reading frames largely maintain similar nucleotide composition at each codon position and amino acid usage, while derived reading frames are significantly different. Our results also indicated that prior to acquisition of a new reading frame, the suppression of in-frame stop codons in the ancestral state is mainly achieved by one-step transition substitutions at the first or second codon position. Finally, the selective forces imposed on these dual coding regions will also be discussed.
Figures

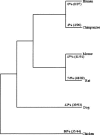

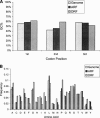

References
-
- Croft, L., Schandorff, S., Clark, F., Burrage, K., Arctander, P., and Mattick, J.S. 2000. ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nat. Genet. 24 340-341. - PubMed
-
- Eigen, M. and Schuster, P. 1979. The hypercycle: A principle of natural self-organzation. Springer-Verlag, Berlin.
-
- Gattiker, A., Gasteiger, E., and Bairoch, A. 2002. ScanProsite: A reference implementation of a PROSITE scanning tool. Appl. Bioinformatics 1 107-108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
