Transposon-free regions in mammalian genomes
- PMID: 16365385
- PMCID: PMC1361711
- DOI: 10.1101/gr.4624306
Transposon-free regions in mammalian genomes
Abstract
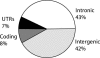
Despite the presence of over 3 million transposons separated on average by approximately 500 bp, the human and mouse genomes each contain almost 1000 transposon-free regions (TFRs) over 10 kb in length. The majority of human TFRs correlate with orthologous TFRs in the mouse, despite the fact that most transposons are lineage specific. Many human TFRs also overlap with orthologous TFRs in the marsupial opossum, indicating that these regions have remained refractory to transposon insertion for long evolutionary periods. Over 90% of the bases covered by TFRs are noncoding, much of which is not highly conserved. Most TFRs are not associated with unusual nucleotide composition, but are significantly associated with genes encoding developmental regulators, suggesting that they represent extended regions of regulatory information that are largely unable to tolerate insertions, a conclusion difficult to reconcile with current conceptions of gene regulation.
Figures

References
-
- Amarger, V., Nguyen, M., Laere, A.S., Braunschweig, M., Nezer, C., Georges, M., and Andersson, L. 2002. Comparative sequence analysis of the INS-IGF2-H19 gene cluster in pigs. Mamm. Genome 13 388-398. - PubMed
-
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S., and Haussler, D. 2004. Ultraconserved elements in the human genome. Science 304 1321-1325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
