Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae
- PMID: 16368004
- PMCID: PMC1343592
- DOI: 10.1186/1471-2148-5-73
Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae
Abstract
Background: Rates of synonymous nucleotide substitutions are, in general, exceptionally low in plant mitochondrial genomes, several times lower than in chloroplast genomes, 10-20 times lower than in plant nuclear genomes, and 50-100 times lower than in many animal mitochondrial genomes. Several cases of moderate variation in mitochondrial substitution rates have been reported in plants, but these mostly involve correlated changes in chloroplast and/or nuclear substitution rates and are therefore thought to reflect whole-organism forces rather than ones impinging directly on the mitochondrial mutation rate. Only a single case of extensive, mitochondrial-specific rate changes has been described, in the angiosperm genus Plantago.
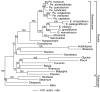
Results: We explored a second potential case of highly accelerated mitochondrial sequence evolution in plants. This case was first suggested by relatively poor hybridization of mitochondrial gene probes to DNA of Pelargonium hortorum (the common geranium). We found that all eight mitochondrial genes sequenced from P. hortorum are exceptionally divergent, whereas chloroplast and nuclear divergence is unexceptional in P. hortorum. Two mitochondrial genes were sequenced from a broad range of taxa of variable relatedness to P. hortorum, and absolute rates of mitochondrial synonymous substitutions were calculated on each branch of a phylogenetic tree of these taxa. We infer one major, approximately 10-fold increase in the mitochondrial synonymous substitution rate at the base of the Pelargonium family Geraniaceae, and a subsequent approximately 10-fold rate increase early in the evolution of Pelargonium. We also infer several moderate to major rate decreases following these initial rate increases, such that the mitochondrial substitution rate has returned to normally low levels in many members of the Geraniaceae. Finally, we find unusually little RNA editing of Geraniaceae mitochondrial genes, suggesting high levels of retroprocessing in their history.
Conclusion: The existence of major, mitochondrial-specific changes in rates of synonymous substitutions in the Geraniaceae implies major and reversible underlying changes in the mitochondrial mutation rate in this family. Together with the recent report of a similar pattern of rate heterogeneity in Plantago, these findings indicate that the mitochondrial mutation rate is a more plastic character in plants than previously realized. Many molecular factors could be responsible for these dramatic changes in the mitochondrial mutation rate, including nuclear gene mutations affecting the fidelity and efficacy of mitochondrial DNA replication and/or repair and--consistent with the lack of RNA editing--exceptionally high levels of "mutagenic" retroprocessing. That the mitochondrial mutation rate has returned to normally low levels in many Geraniaceae raises the possibility that, akin to the ephemerality of mutator strains in bacteria, selection favors a low mutation rate in plant mitochondria.
Figures



References
-
- Wolfe KH. Molecular evolution of plants: more genomes, fewer generalities. In: Andersson B, Salter AH, Barber J, editor. Molecular Genetics of Photosynthesis. Oxford: IRL Press; 1996. pp. 45–57.
-
- Gaut BS. Molecular clocks and nucleotide substitution rates in higher plants. In: Hecht MK, MacIntyre RJ, Clegg MT, editor. Evolutionary Biology. Vol. 30. New York: Plenum Press; 1998. pp. 93–120.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

