Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa
- PMID: 16368012
- PMCID: PMC1368998
- DOI: 10.1186/1471-2148-5-72
Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa
Abstract
Background: Calcium signaling plays a prominent role in plants for coordinating a wide range of developmental processes and responses to environmental cues. Stimulus-specific generation of intracellular calcium transients, decoding of calcium signatures, and transformation of the signal into cellular responses are integral modules of the transduction process. Several hundred proteins with functions in calcium signaling circuits have been identified, and the number of downstream targets of calcium sensors is expected to increase. We previously identified a novel, calmodulin-binding nuclear protein, IQD1, which stimulates glucosinolate accumulation and plant defense in Arabidopsis thaliana. Here, we present a comparative genome-wide analysis of a new class of putative calmodulin target proteins in Arabidopsis and rice.
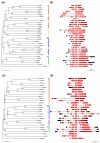
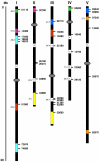
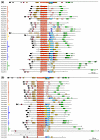
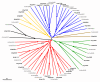
Results: We identified and analyzed 33 and 29 IQD1-like genes in Arabidopsis thaliana and Oryza sativa, respectively. The encoded IQD proteins contain a plant-specific domain of 67 conserved amino acid residues, referred to as the IQ67 domain, which is characterized by a unique and repetitive arrangement of three different calmodulin recruitment motifs, known as the IQ, 1-5-10, and 1-8-14 motifs. We demonstrated calmodulin binding for IQD20, the smallest IQD protein in Arabidopsis, which consists of a C-terminal IQ67 domain and a short N-terminal extension. A striking feature of IQD proteins is the high isoelectric point (approximately 10.3) and frequency of serine residues (approximately 11%). We compared the Arabidopsis and rice IQD gene families in terms of gene structure, chromosome location, predicted protein properties and motifs, phylogenetic relationships, and evolutionary history. The existence of an IQD-like gene in bryophytes suggests that IQD proteins are an ancient family of calmodulin-binding proteins and arose during the early evolution of land plants.
Conclusion: Comparative phylogenetic analyses indicate that the major IQD gene lineages originated before the monocot-eudicot divergence. The extant IQD loci in Arabidopsis primarily resulted from segmental duplication and reflect preferential retention of paralogous genes, which is characteristic for proteins with regulatory functions. Interaction of IQD1 and IQD20 with calmodulin and the presence of predicted calmodulin binding sites in all IQD family members suggest that IQD proteins are a new class of calmodulin targets. The basic isoelectric point of IQD proteins and their frequently predicted nuclear localization suggest that IQD proteins link calcium signaling pathways to the regulation of gene expression. Our comparative genomics analysis of IQD genes and encoded proteins in two model plant species provides the first step towards the functional dissection of this emerging family of putative calmodulin targets.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

