Inhibition of chlamydiae by primary alcohols correlates with the strain-specific complement of plasticity zone phospholipase D genes
- PMID: 16368959
- PMCID: PMC1346656
- DOI: 10.1128/IAI.74.1.73-80.2006
Inhibition of chlamydiae by primary alcohols correlates with the strain-specific complement of plasticity zone phospholipase D genes
Abstract
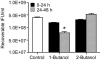
Members of the genus Chlamydia are obligate intracellular pathogens that have a unique biphasic developmental cycle and interactions with host cells. Many genes that dictate host infection tropism and, putatively, pathogenic manifestations of disease are clustered in a hypervariable region of the genome termed the plasticity zone (PZ). Comparative genomics studies have determined that an uncharacterized family of PZ genes encoding orthologs of eukaryotic and prokaryotic members of the phospholipase D (PLD) enzyme family varies among chlamydiae. Here, we show that the PZ PLD (pzPLD) of Chlamydia trachomatis are transcribed during both normal and persistent infection and that the corresponding PLD proteins are predominantly localized in reticulate bodies on the inner leaflet of the inclusion membrane. Further, we show that strains of chlamydiae encoding the pzPLD, but not a strain lacking these genes, are inhibited by primary alcohols, potent PLD inhibitors, during growth in HeLa 229 cells. This inhibitory effect is amplified approximately 5,000-fold during recovery from persistent infection. These findings suggest that the chlamydial pzPLD may be important, strain-specific, pathogenesis factors in vivo.
Figures





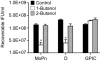

References
-
- Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Investig. 111:1757-1769. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

