Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis
- PMID: 16368989
- PMCID: PMC1346599
- DOI: 10.1128/IAI.74.1.340-351.2006
Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis
Abstract
Entamoeba histolytica is a protozoan parasite that causes colitis and liver abscesses. Several Entamoeba species and strains with differing levels of virulence have been identified. E. histolytica HM-1:IMSS is a virulent strain, E. histolytica Rahman is a nonvirulent strain, and Entamoeba dispar is a nonvirulent species. We used an E. histolytica DNA microarray consisting of 2,110 genes to assess the transcriptional differences between these species/strains with the goal of identifying genes whose expression correlated with a virulence phenotype. We found 415 genes expressed at lower levels in E. dispar and 32 genes with lower expression in E. histolytica Rahman than in E. histolytica HM-1:IMSS. Overall, 29 genes had decreased expression in both the nonvirulent species/strains than the virulent E. histolytica HM-1:IMSS. Interestingly, a number of genes with potential roles in stress response and virulence had decreased expression in either one or both nonvirulent Entamoeba species/strains. These included genes encoding Fe hydrogenase (9.m00419), peroxiredoxin (176.m00112), type A flavoprotein (6.m00467), lysozyme (6.m00454), sphingomyelinase C (29.m00231), and a hypothetical protein with homology to both a Plasmodium sporozoite threonine-asparagine-rich protein (STARP) and a streptococcal hemagglutinin (238.m00054). The function of these genes in Entamoeba and their specific roles in parasite virulence need to be determined. We also found that a number of the non-long-terminal-repeat retrotransposons (EhLINEs and EhSINEs), which have been shown to modulate gene expression and genomic evolution, had lower expression in the nonvirulent species/strains than in E. histolytica HM-1:IMSS. Our results, identifying expression profiles and patterns indicative of a virulence phenotype, may be useful in characterizing the transcriptional framework of virulence.
Figures
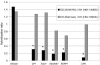
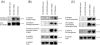


 , >85% nucleotide identity; □, <80% nucleotide identity). No high-homology hit was found for ORF1 in E. dispar SAW760; however, a 1,600-bp region encompassing the reverse transcriptase domain showed 85% identity in E. dispar SAW760. (B) The average expression levels of EhLINE and EhSINE in E. histolytica Rahman and E. dispar SAW760 relative to those in E. histolytica HM-1:IMSS are shown. For each element, 50 clones with the highest similarity to the consensus sequence for each EhLINE or EhSINE were used. The copy number (adapted from Bakre et al. [5]), median BLASTN E-value for the 50 clones with highest homology, and genomic sequence similarity are also displayed. Expression levels that are significantly different from that of E. histolytica HM-1:IMSS are labeled by an asterisk and denote a P value of <0.05.
, >85% nucleotide identity; □, <80% nucleotide identity). No high-homology hit was found for ORF1 in E. dispar SAW760; however, a 1,600-bp region encompassing the reverse transcriptase domain showed 85% identity in E. dispar SAW760. (B) The average expression levels of EhLINE and EhSINE in E. histolytica Rahman and E. dispar SAW760 relative to those in E. histolytica HM-1:IMSS are shown. For each element, 50 clones with the highest similarity to the consensus sequence for each EhLINE or EhSINE were used. The copy number (adapted from Bakre et al. [5]), median BLASTN E-value for the 50 clones with highest homology, and genomic sequence similarity are also displayed. Expression levels that are significantly different from that of E. histolytica HM-1:IMSS are labeled by an asterisk and denote a P value of <0.05.
 , >85% nucleotide identity;
, >85% nucleotide identity;  , >80% nucleotide identity; □, <80% nucleotide identity). ×, stop codons; ▵, mutated stop or start codons.
, >80% nucleotide identity; □, <80% nucleotide identity). ×, stop codons; ▵, mutated stop or start codons.References
-
- Akbar, M. A., N. S. Chatterjee, P. Sen, A. Debnath, A. Pal, T. Bera, and P. Das. 2004. Genes induced by a high-oxygen environment in Entamoeba histolytica. Mol. Biochem. Parasitol. 133:187-196. - PubMed
-
- Ankri, S., F. Padilla-Vaca, T. Stolarsky, L. Koole, U. Katz, and D. Mirelman. 1999. Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNac lectin complex inhibits Entamoeba histolytica virulence. Mol. Microbiol. 33:327-337. - PubMed
-
- Ankri, S., T. Stolarsky, and D. Mirelman. 1998. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol. Microbiol. 28:777-785. - PubMed
-
- Bakre, A. A., K. Rawal, R. Ramaswamy, A. Bhattacharya, and S. Bhattacharya. 2005. The LINEs and SINEs of Entamoeba histolytica: comparative analysis and genomic distribution. Exp. Parasitol. 110:207-213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

