Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection
- PMID: 16369534
- PMCID: PMC7097088
- DOI: 10.1038/ng1698
Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection
Abstract
Severe acute respiratory syndrome (SARS) is caused by infection of a previously undescribed coronavirus (CoV). L-SIGN, encoded by CLEC4M (also known as CD209L), is a SARS-CoV binding receptor that has polymorphism in its extracellular neck region encoded by the tandem repeat domain in exon 4. Our genetic risk association study shows that individuals homozygous for CLEC4M tandem repeats are less susceptible to SARS infection. L-SIGN is expressed in both non-SARS and SARS-CoV-infected lung. Compared with cells heterozygous for L-SIGN, cells homozygous for L-SIGN show higher binding capacity for SARS-CoV, higher proteasome-dependent viral degradation and a lower capacity for trans infection. Thus, homozygosity for L-SIGN plays a protective role during SARS infection.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
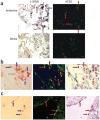

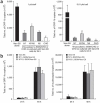
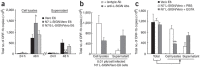

Comment in
-
Lack of support for an association between CLEC4M homozygosity and protection against SARS coronavirus infection.Nat Genet. 2007 Jun;39(6):691-2; author reply 694-6. doi: 10.1038/ng0607-691. Nat Genet. 2007. PMID: 17534354 Free PMC article. No abstract available.
-
Lack of support for an association between CLEC4M homozygosity and protection against SARS coronavirus infection.Nat Genet. 2007 Jun;39(6):692-4; author reply 694-6. doi: 10.1038/ng0607-692. Nat Genet. 2007. PMID: 17534355 Free PMC article. No abstract available.
Similar articles
-
Lack of support for an association between CLEC4M homozygosity and protection against SARS coronavirus infection.Nat Genet. 2007 Jun;39(6):692-4; author reply 694-6. doi: 10.1038/ng0607-692. Nat Genet. 2007. PMID: 17534355 Free PMC article. No abstract available.
-
Lack of support for an association between CLEC4M homozygosity and protection against SARS coronavirus infection.Nat Genet. 2007 Jun;39(6):691-2; author reply 694-6. doi: 10.1038/ng0607-691. Nat Genet. 2007. PMID: 17534354 Free PMC article. No abstract available.
-
Role of polymorphisms of the inflammatory response genes and DC-SIGNR in genetic susceptibility to SARS and other infections.Hong Kong Med J. 2008 Aug;14 Suppl 4:31-5. Hong Kong Med J. 2008. PMID: 18708672
-
DC-SIGN and L-SIGN: the SIGNs for infection.J Mol Med (Berl). 2008 Aug;86(8):861-74. doi: 10.1007/s00109-008-0350-2. Epub 2008 May 6. J Mol Med (Berl). 2008. PMID: 18458800 Free PMC article. Review.
-
Pathogenesis of severe acute respiratory syndrome.Curr Opin Immunol. 2005 Aug;17(4):404-10. doi: 10.1016/j.coi.2005.05.009. Curr Opin Immunol. 2005. PMID: 15950449 Free PMC article. Review.
Cited by
-
Coronavirus: Pure Infectious Disease or Genetic Predisposition.Adv Exp Med Biol. 2021;1318:91-107. doi: 10.1007/978-3-030-63761-3_6. Adv Exp Med Biol. 2021. PMID: 33973174
-
Unprecedented selectivity for homologous lectin targets: differential targeting of the viral receptors L-SIGN and DC-SIGN.Chem Sci. 2024 Aug 27;15(37):15352-66. doi: 10.1039/d4sc02980a. Online ahead of print. Chem Sci. 2024. PMID: 39246372 Free PMC article.
-
C-type lectin receptors in tuberculosis: what we know.Med Microbiol Immunol. 2016 Dec;205(6):513-535. doi: 10.1007/s00430-016-0470-1. Epub 2016 Jul 28. Med Microbiol Immunol. 2016. PMID: 27469378 Review.
-
Lack of support for an association between CLEC4M homozygosity and protection against SARS coronavirus infection.Nat Genet. 2007 Jun;39(6):692-4; author reply 694-6. doi: 10.1038/ng0607-692. Nat Genet. 2007. PMID: 17534355 Free PMC article. No abstract available.
-
Cholesterol-Rich Lipid Rafts as Platforms for SARS-CoV-2 Entry.Front Immunol. 2021 Dec 16;12:796855. doi: 10.3389/fimmu.2021.796855. eCollection 2021. Front Immunol. 2021. PMID: 34975904 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

