Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation
- PMID: 16373480
- PMCID: PMC1370902
- DOI: 10.1261/rna.2153206
Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation
Abstract
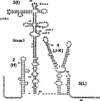
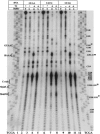
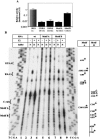
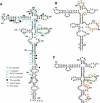
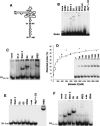
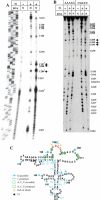
Internal ribosome entry site (IRES) elements consist of highly structured RNA regions that determine internal initiation of translation. We have previously shown that the foot-and-mouth disease virus (FMDV) IRES contains a GNRA tetraloop spanning residues G178UAA181. Here we show that tertiary RNA interactions dependent on the GNRA motif determine the structural organization of the central domain. By using mutational analysis in combination with RNA probing, we have identified distant reciprocal interactions between the GNRA motif and the invariant region G240CACG244, termed motif A. Mutations in motif A caused a decrease in IRES activity as severe as the GUAG substitution in the GNRA motif. Substitutions in either GNRA or motif A sequences induced a common reorganization around the conserved R199AAA202 stem-loop, suggesting that the latter contributes to stabilize the GNRA-motif A interaction. This finding was also consistent with a significant increase in the efficiency of RNA-RNA interactions determined in gel shift assays using as probe the hairpin that contains the GNRA motif compared to a transcript encompassing the entire apical region of the central domain. Thus, we propose that the central domain of the FMDV IRES contains a structural conformation essential for IRES activity stabilized by a tertiary contact involving residues in the GNRA tetraloop and motif A conserved sequences.
Figures

Similar articles
-
Structural organization of a viral IRES depends on the integrity of the GNRA motif.RNA. 2003 Nov;9(11):1333-44. doi: 10.1261/rna.5950603. RNA. 2003. PMID: 14561883 Free PMC article.
-
Characterization of a cyanobacterial RNase P ribozyme recognition motif in the IRES of foot-and-mouth disease virus reveals a unique structural element.RNA. 2007 Jun;13(6):849-59. doi: 10.1261/rna.506607. Epub 2007 Apr 20. RNA. 2007. PMID: 17449727 Free PMC article.
-
Structural analysis provides insights into the modular organization of picornavirus IRES.Virology. 2011 Jan 20;409(2):251-61. doi: 10.1016/j.virol.2010.10.013. Epub 2010 Nov 5. Virology. 2011. PMID: 21056890
-
An atypical IRES within the 5' UTR of a dicistrovirus genome.Virus Res. 2009 Feb;139(2):157-65. doi: 10.1016/j.virusres.2008.07.017. Epub 2008 Sep 11. Virus Res. 2009. PMID: 18755228 Review.
-
An RNA folding motif: GNRA tetraloop-receptor interactions.Q Rev Biophys. 2013 Aug;46(3):223-64. doi: 10.1017/S0033583513000048. Epub 2013 Aug 5. Q Rev Biophys. 2013. PMID: 23915736 Review.
Cited by
-
Analysis of the interaction between host factor Sam68 and viral elements during foot-and-mouth disease virus infections.Virol J. 2015 Dec 23;12:224. doi: 10.1186/s12985-015-0452-8. Virol J. 2015. PMID: 26695943 Free PMC article.
-
In vitro molecular characterization of RNA-proteins interactions during initiation of translation of a wild-type and a mutant Coxsackievirus B3 RNAs.Mol Biotechnol. 2013 Jun;54(2):515-27. doi: 10.1007/s12033-012-9592-x. Mol Biotechnol. 2013. PMID: 22923320
-
Deconstructing internal ribosome entry site elements: an update of structural motifs and functional divergences.Open Biol. 2018 Nov 28;8(11):180155. doi: 10.1098/rsob.180155. Open Biol. 2018. PMID: 30487301 Free PMC article. Review.
-
Modifying inter-cistronic sequence significantly enhances IRES dependent second gene expression in bicistronic vector: Construction of optimised cassette for gene therapy of familial hypercholesterolemia.Noncoding RNA Res. 2018 Nov 22;4(1):1-14. doi: 10.1016/j.ncrna.2018.11.005. eCollection 2019 Mar. Noncoding RNA Res. 2018. PMID: 30891532 Free PMC article.
-
Host-like RNA Elements Regulate Virus Translation.Viruses. 2024 Mar 20;16(3):468. doi: 10.3390/v16030468. Viruses. 2024. PMID: 38543832 Free PMC article. Review.
References
-
- Aldaz-Carroll, L., Tallet, B., Dausse, E., Yurchenko, L., and Toulmé, J.-J. 2002. Apical loop-internal loop interactions: A new RNA–RNA recognition motif identified through in vitro selection against RNA hairpins of the hepatitis C virus mRNA. Biochemistry 41: 5883–5893. - PubMed
-
- Belsham, G.J. and Jackson, R.J. 2000. Translation initiation on picornavirus RNA. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 869–900. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Branch, A.D., Benenfeld, B.J., and Robertson, H.D. 1989. RNA fingerprinting. Methods Enzymol. 180: 130–154. - PubMed
-
- Brunel, C. and Romby, P. 2000. Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol. 318: 3–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
