Prp8p dissection reveals domain structure and protein interaction sites
- PMID: 16373487
- PMCID: PMC1370899
- DOI: 10.1261/rna.2281306
Prp8p dissection reveals domain structure and protein interaction sites
Abstract
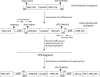
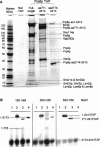
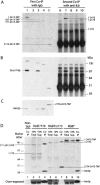
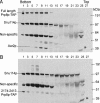
We describe a novel approach to characterize the functional domains of a protein in vivo. This involves the use of a custom-built Tn5-based transposon that causes the expression of a target gene as two contiguous polypeptides. When used as a genetic screen to dissect the budding yeast PRP8 gene, this showed that Prp8 protein could be dissected into three distinct pairs of functional polypeptides. Thus, four functional domains can be defined in the 2413-residue Prp8 protein, with boundaries in the regions of amino acids 394-443, 770, and 2170-2179. The central region of the protein was resistant to dissection by this approach, suggesting that it represents one large functional unit. The dissected constructs allowed investigation of factors that associate strongly with the N- or the C-terminal Prp8 protein fragments. Thus, the U5 snRNP protein Snu114p associates with Prp8p in the region 437-770, whereas fragmenting Prp8p at residue 2173 destabilizes its association with Aar2p.
Figures

Similar articles
-
prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast.Nat Struct Mol Biol. 2007 Nov;14(11):1077-83. doi: 10.1038/nsmb1303. Epub 2007 Oct 14. Nat Struct Mol Biol. 2007. PMID: 17934474 Free PMC article.
-
Structural basis for dual roles of Aar2p in U5 snRNP assembly.Genes Dev. 2013 Mar 1;27(5):525-40. doi: 10.1101/gad.213207.113. Epub 2013 Feb 26. Genes Dev. 2013. PMID: 23442228 Free PMC article.
-
Mechanism for Aar2p function as a U5 snRNP assembly factor.Genes Dev. 2011 Aug 1;25(15):1601-12. doi: 10.1101/gad.635911. Epub 2011 Jul 15. Genes Dev. 2011. PMID: 21764848 Free PMC article.
-
The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p.RNA. 2001 Nov;7(11):1554-65. RNA. 2001. PMID: 11720285 Free PMC article.
-
Structural dynamics of the N-terminal domain and the Switch loop of Prp8 during spliceosome assembly and activation.Nucleic Acids Res. 2018 May 4;46(8):3833-3840. doi: 10.1093/nar/gky242. Nucleic Acids Res. 2018. PMID: 29635373 Free PMC article. Review.
Cited by
-
prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast.Nat Struct Mol Biol. 2007 Nov;14(11):1077-83. doi: 10.1038/nsmb1303. Epub 2007 Oct 14. Nat Struct Mol Biol. 2007. PMID: 17934474 Free PMC article.
-
Functional roles of protein splicing factors.Biosci Rep. 2012 Aug;32(4):345-59. doi: 10.1042/BSR20120007. Biosci Rep. 2012. PMID: 22762203 Free PMC article. Review.
-
Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants.Protein Sci. 2007 Jun;16(6):1024-31. doi: 10.1110/ps.072872007. Epub 2007 May 1. Protein Sci. 2007. PMID: 17473007 Free PMC article.
-
A snRNP's ordered path to maturity.Genes Dev. 2011 Aug 1;25(15):1563-7. doi: 10.1101/gad.17311211. Genes Dev. 2011. PMID: 21828266 Free PMC article.
-
The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine.Proteomics. 2010 Nov;10(22):4128-41. doi: 10.1002/pmic.201000354. Epub 2010 Nov 2. Proteomics. 2010. PMID: 21080498 Free PMC article. Review.
References
-
- Achsel, T., Ahrens, K., Brahms, H., Teigelkamp, S., and Lührmann, R. 1998. The human U5–220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18: 6756–6766. - PMC - PubMed
-
- Bartels, C., Urlaub, H., Luhrmann, R., and Fabrizio, P. 2003. Muta-genesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 278: 28324–28334. - PubMed
-
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. - PubMed
-
- Doolittle, R.F. 1995. The multiplicity of domains in proteins. Annu. Rev. Biochem. 64: 287–314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
