A computational screen for mammalian pseudouridylation guide H/ACA RNAs
- PMID: 16373490
- PMCID: PMC1370881
- DOI: 10.1261/rna.2210406
A computational screen for mammalian pseudouridylation guide H/ACA RNAs
Abstract
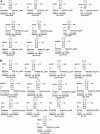
The box H/ACA RNA gene family is one of the largest non-protein-coding gene families in eukaryotes and archaea. Recently, we developed snoGPS, a computational screening program for H/ACA snoRNAs, and applied it to Saccharomyces cerevisiae. We report here results of extending our method to screen for H/ACA RNAs in multiple large genomes of related species, and apply it to the human, mouse, and rat genomes. Because of the 250-fold larger search space compared to S. cerevisiae, significant enhancements to our algorithms were required. Complementing extensive cloning experiments performed by others, our findings include the detection and experimental verification of seven new mammalian H/ACA RNAs and the prediction of 23 new H/ACA RNA pseudouridine guide assignments. These assignments include four for H/ACA RNAs previously classified as orphan H/ACA RNAs with no known targets. We also determined systematic syntenic conservation among human and mouse H/ACA RNAs. With this work, 82 of 97 ribosomal RNA pseudouridines and 18 of 32 spliceosomal RNA pseudouridines in mammals have been linked to H/ACA guide RNAs.
Figures


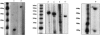

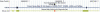
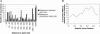
References
-
- Bachellerie, J.P., Cavaille, J., and Huttenhofer, A. 2002. The expanding snoRNA world. Biochimie 84: 775–790. - PubMed
-
- Bertrand, E. and Fournier, M.J. 2004. The snoRNPs and related machines: Ancient devices that mediate maturation of rRNA and other RNAs. In The nucleolus (ed. M. Olson), pp. 225–261. Landes Bioscience, Georgetown, TX.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
