The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling
- PMID: 16374511
- PMCID: PMC1369247
- DOI: 10.1038/sj.embor.7400593
The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling
Abstract
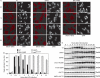
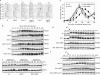
Double-strand breaks (DSBs) elicit a DNA damage response, resulting in checkpoint-mediated cell-cycle delay and DNA repair. The Saccharomyces cerevisiae Sae2 protein is known to act together with the MRX complex in meiotic DSB processing, as well as in DNA damage response during the mitotic cell cycle. Here, we report that cells lacking Sae2 fail to turn off both Mec1- and Tel1-dependent checkpoints activated by a single irreparable DSB, and delay Mre11 foci disassembly at DNA breaks, indicating that Sae2 may negatively regulate checkpoint signalling by modulating MRX association at damaged DNA. Consistently, high levels of Sae2 prevent checkpoint activation and impair MRX foci formation in response to unrepaired DSBs. Mec1- and Tel1-dependent Sae2 phosphorylation is necessary for these Sae2 functions, suggesting that the two kinases, once activated, may regulate checkpoint switch off through Sae2-mediated inhibition of MRX signalling.
Figures


References
-
- Clerici M, Mantiero D, Lucchini G, Longhese MP (2005) The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double-strand break ends. J Biol Chem 280: 38631–38638 - PubMed
-
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE (1998) Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

