Evolutionary programming as a platform for in silico metabolic engineering
- PMID: 16375763
- PMCID: PMC1327682
- DOI: 10.1186/1471-2105-6-308
Evolutionary programming as a platform for in silico metabolic engineering
Abstract
Background: Through genetic engineering it is possible to introduce targeted genetic changes and hereby engineer the metabolism of microbial cells with the objective to obtain desirable phenotypes. However, owing to the complexity of metabolic networks, both in terms of structure and regulation, it is often difficult to predict the effects of genetic modifications on the resulting phenotype. Recently genome-scale metabolic models have been compiled for several different microorganisms where structural and stoichiometric complexity is inherently accounted for. New algorithms are being developed by using genome-scale metabolic models that enable identification of gene knockout strategies for obtaining improved phenotypes. However, the problem of finding optimal gene deletion strategy is combinatorial and consequently the computational time increases exponentially with the size of the problem, and it is therefore interesting to develop new faster algorithms.
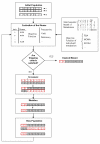
Results: In this study we report an evolutionary programming based method to rapidly identify gene deletion strategies for optimization of a desired phenotypic objective function. We illustrate the proposed method for two important design parameters in industrial fermentations, one linear and other non-linear, by using a genome-scale model of the yeast Saccharomyces cerevisiae. Potential metabolic engineering targets for improved production of succinic acid, glycerol and vanillin are identified and underlying flux changes for the predicted mutants are discussed.
Conclusion: We show that evolutionary programming enables solving large gene knockout problems in relatively short computational time. The proposed algorithm also allows the optimization of non-linear objective functions or incorporation of non-linear constraints and additionally provides a family of close to optimal solutions. The identified metabolic engineering strategies suggest that non-intuitive genetic modifications span several different pathways and may be necessary for solving challenging metabolic engineering problems.
Figures

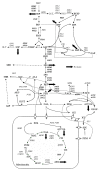
 style indicates a lumped pathway. Multiple names for a reaction indicate the presence of iso-enzymes. The nomenclature of the metabolites can be found in the Supplementary table 1 [see Additional file 1]. The figure is partially adapted from Forster et al. (2002) [32].
style indicates a lumped pathway. Multiple names for a reaction indicate the presence of iso-enzymes. The nomenclature of the metabolites can be found in the Supplementary table 1 [see Additional file 1]. The figure is partially adapted from Forster et al. (2002) [32].
References
-
- Nielsen J. Metabolic Engineering. Applied Microbiology and Biotechnology. 2001;55:263–283. doi: 10.1007/s002530000511. http://www.springerlink.com/openurl.asp?genre=article&issn=0175-7598&vol... - DOI - PubMed
-
- Stephanopoulos G, Aristidou AA, Nielsen J. Metabolic engineering Principles and methodologies. 1. San Diego, Academic Press; 1998.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

