Solution structure of psi32-modified anticodon stem-loop of Escherichia coli tRNAPhe
- PMID: 16377777
- PMCID: PMC1322268
- DOI: 10.1093/nar/gki1004
Solution structure of psi32-modified anticodon stem-loop of Escherichia coli tRNAPhe
Abstract
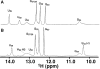
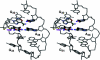
Nucleoside base modifications can alter the structures and dynamics of RNA molecules and are important in tRNAs for maintaining translational fidelity and efficiency. The unmodified anticodon stem-loop from Escherichia coli tRNA(Phe) forms a trinucleotide loop in solution, but Mg2+ and dimethylallyl modification of A37 N6 destabilize the loop-proximal base pairs and increase the mobility of the loop nucleotides. The anticodon arm has three additional modifications, psi32, psi39, and A37 C2-thiomethyl. We have used NMR spectroscopy to investigate the structural and dynamical effects of psi32 on the anticodon stem-loop from E.coli tRNA(Phe). The psi32 modification does not significantly alter the structure of the anticodon stem-loop relative to the unmodified parent molecule. The stem of the RNA molecule includes base pairs psi32-A38 and U33-A37 and the base of psi32 stacks between U33 and A31. The glycosidic bond of psi32 is in the anti configuration and is paired with A38 in a Watson-Crick geometry, unlike residue 32 in most crystal structures of tRNA. The psi32 modification increases the melting temperature of the stem by approximately 3.5 degrees C, although the psi32 and U33 imino resonances are exchange broadened. The results suggest that psi32 functions to preserve the stem integrity in the presence of additional loop modifications or after reorganization of the loop into a translationally functional conformation.
Figures
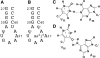




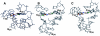
Similar articles
-
Base pairing within the psi32,psi39-modified anticodon arm of Escherichia coli tRNA(Phe).J Am Chem Soc. 2006 Dec 13;128(49):15570-1. doi: 10.1021/ja0659368. J Am Chem Soc. 2006. PMID: 17147349
-
Metal ion stabilization of the U-turn of the A37 N6-dimethylallyl-modified anticodon stem-loop of Escherichia coli tRNAPhe.Biochemistry. 2004 Jan 13;43(1):55-66. doi: 10.1021/bi0353676. Biochemistry. 2004. PMID: 14705931
-
Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNA(Phe).J Mol Biol. 2002 Jun 21;319(5):1015-34. doi: 10.1016/S0022-2836(02)00382-0. J Mol Biol. 2002. PMID: 12079344
-
The effect of pseudouridine and pH on the structure and dynamics of the anticodon stem-loop of tRNA(Lys,3).Nucleic Acids Symp Ser. 1997;(36):56-7. Nucleic Acids Symp Ser. 1997. PMID: 9478205 Review.
-
Loop stereochemistry and dynamics in transfer RNA.J Biomol Struct Dyn. 1983 Oct;1(2):337-55. doi: 10.1080/07391102.1983.10507446. J Biomol Struct Dyn. 1983. PMID: 6401114 Review.
Cited by
-
Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.Nucleic Acids Res. 2020 Sep 25;48(17):9918-9930. doi: 10.1093/nar/gkaa667. Nucleic Acids Res. 2020. PMID: 32785618 Free PMC article.
-
Mining for Perchlorate Resistance Genes in Microorganisms From Sediments of a Hypersaline Pond in Atacama Desert, Chile.Front Microbiol. 2021 Jul 23;12:723874. doi: 10.3389/fmicb.2021.723874. eCollection 2021. Front Microbiol. 2021. PMID: 34367123 Free PMC article.
-
Conformation effects of base modification on the anticodon stem-loop of Bacillus subtilis tRNA(Tyr).J Mol Biol. 2011 Sep 16;412(2):285-303. doi: 10.1016/j.jmb.2011.07.010. Epub 2011 Jul 19. J Mol Biol. 2011. PMID: 21782828 Free PMC article.
-
The crystal structure of unmodified tRNAPhe from Escherichia coli.Nucleic Acids Res. 2010 Jul;38(12):4154-62. doi: 10.1093/nar/gkq133. Epub 2010 Mar 4. Nucleic Acids Res. 2010. PMID: 20203084 Free PMC article.
-
A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.J Mol Biol. 2020 Feb 14;432(4):913-929. doi: 10.1016/j.jmb.2019.12.016. Epub 2020 Jan 14. J Mol Biol. 2020. PMID: 31945376 Free PMC article.
References
-
- Björk G.R. Biosynthesis and function of modified nucleosides. In: Söll D., RajBhandary U.L., editors. tRNA Structure, Biosynthesis, and Function. Washington, D.C.: American Society for Microbiology Press; 1995. pp. 165–206.
-
- Garcia G.A., Goodenough-Lashua D.M. Mechanisms of modifying enzymes. In: Grosjean H., Benne R., editors. Modification and Editing of RNA: The Alteration of RNA Structure and Function. Washington, D.C.: ASM Press; 1998. pp. 135–168.
-
- Winkler M.E. Genetics and regulation of tRNA and rRNA modification. In: Grosjean H., Benne R., editors. Modification and Editing of RNA: The Alteration of RNA Structure and Function. Washington, D.C.: ASM Press; 1998. pp. 441–469.
-
- Agris P.F. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 1996;53:79–129. - PubMed
-
- Charette M., Gray M.W. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

