Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication
- PMID: 16378999
- PMCID: PMC1346866
- DOI: 10.1128/JVI.80.2.975-984.2006
Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication
Abstract
A total of 47 clinical samples were identified during an active surveillance program of respiratory infections in Buenos Aires (BA) (1999 to 2004) that contained sequences of human respiratory syncytial virus (HRSV) with a 60-nucleotide duplication in the attachment (G) protein gene. This duplication was analogous to that previously described for other three viruses also isolated in Buenos Aires in 1999 (A. Trento et al., J. Gen. Virol. 84:3115-3120, 2003). Phylogenetic analysis indicated that BA sequences with that duplication shared a common ancestor (dated about 1998) with other HRSV G sequences reported worldwide after 1999. The duplicated nucleotide sequence was an exact copy of the preceding 60 nucleotides in early viruses, but both copies of the duplicated segment accumulated nucleotide substitutions in more recent viruses at a rate apparently higher than in other regions of the G protein gene. The evolution of the viruses with the duplicated G segment apparently followed the overall evolutionary pattern previously described for HRSV, and this genotype has replaced other prevailing antigenic group B genotypes in Buenos Aires and other places. Thus, the duplicated segment represents a natural tag that can be used to track the dissemination and evolution of HRSV in an unprecedented setting. We have taken advantage of this situation to reexamine the molecular epidemiology of HRSV and to explore the natural history of this important human pathogen.
Figures
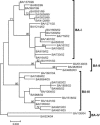
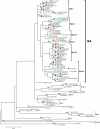

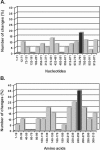
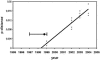
 estimated by the maximum-likelihood method and the 95% confidence interval.
estimated by the maximum-likelihood method and the 95% confidence interval.
References
-
- Anderson, L. J., J. C. Hierholzer, C. Tsou, R. M. Hendry, B. F. Fernie, Y. Stone, and K. McIntosh. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626-633. - PubMed
-
- Blanc, A., A. Delfraro, S. Frabasile, and J. Arbiza. 2005. Genotypes of respiratory syncytial virus group B identified in Uruguay. Arch. Virol. 150:603-609. - PubMed
-
- Cane, P. A. 2001. Molecular epidemiology of respiratory syncytial virus. Rev. Med. Virol. 11:103-116. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

