Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques
- PMID: 16379001
- PMCID: PMC1346845
- DOI: 10.1128/JVI.80.2.999-1014.2006
Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques
Abstract
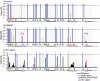
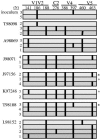
We have analyzed changes to proviral Env gp120 sequences and the development of neutralizing antibodies (NAbs) during 1 year of simian/human immunodeficiency virus SHIV-89.6P infection in 11 Macaca nemestrina macaques. Seven macaques had significant env divergence from that of the inoculum, and macaques with greater divergence had higher titers of homologous NAbs. Substitutions in sequons encoding potential N-linked glycosylation sites (PNGs) were among the first to be established, although overall the total number of sequons did not increase significantly. The majority (19 of 23) of PNGs present in the inoculum were conserved in the sequences from all macaques. Statistically significant variations in PNGs occurred in multiple macaques within constrained regions we term "hot spots," resulting in the selection of sequences more similar to the B consensus. These included additions on V1, the N-terminal side of V4, and the outer region of C2. Complex mutational patterns resulted in convergent PNG shifts in V2 and V5. Charge changes in Env V1V2, resulting in a net acidic charge, and a proline addition in V5 occurred in several macaques. Molecular modeling of the 89.6P sequence showed that the conserved glycans lie on the silent face of Env and that many are proximal to disulfide bonds, while PNG additions and shifts are proximal to the CD4 binding site. Nonsynonymous-to-synonymous substitution ratios suggest that these changes result from selective pressure. This longitudinal and cross-sectional study of mutations in human immunodeficiency virus (HIV) env in the SHIV background provides evidence that there are more constraints on the configuration of the glycan shield than were previously appreciated.
Figures





References
-
- Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. - PubMed
-
- Anonymous. 1997. Lasergene software suite. DNASTAR Inc., Madison, Wis.
-
- Balzarini, J., S. Hatse, K. Vermeire, K. Princen, S. Aquaro, C.-F. Perno, E. D. Clercq, H. Egberink, G. V. Mooter, W. Peumans, E. V. Damme, and D. Schols. 2004. Mannose-specific plant lectins from the Amaryllidaceae family qualify as efficient microbicides for prevention of human immunodeficiency virus infection. Antimicrob. Agents Chemother. 48:3858-3870. - PMC - PubMed
-
- Balzarini, J., K. V. Laethem, S. Hatse, M. Froeyen, E. V. Damme, A. Bolmstedt, W. Peumans, E. D. Clercq, and D. Schols. 2005. Marked depletion of glycosylation sites in HIV-1 gp120 under selection pressure by the mannose-specific plant lectins of Hippeastrum hybrid and Galanthus nivalis. Mol. Pharmacol. 67:1556-1565. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

