Coupling of the nucleus and cytoplasm: role of the LINC complex
- PMID: 16380439
- PMCID: PMC2063530
- DOI: 10.1083/jcb.200509124
Coupling of the nucleus and cytoplasm: role of the LINC complex
Abstract
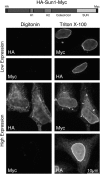
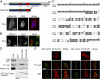
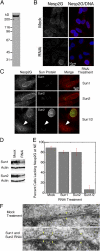
The nuclear envelope defines the barrier between the nucleus and cytoplasm and features inner and outer membranes separated by a perinuclear space (PNS). The inner nuclear membrane contains specific integral proteins that include Sun1 and Sun2. Although the outer nuclear membrane (ONM) is continuous with the endoplasmic reticulum, it is nevertheless enriched in several integral membrane proteins, including nesprin 2 Giant (nesp2G), an 800-kD protein featuring an NH(2)-terminal actin-binding domain. A recent study (Padmakumar, V.C., T. Libotte, W. Lu, H. Zaim, S. Abraham, A.A. Noegel, J. Gotzmann, R. Foisner, and I. Karakesisoglou. 2005. J. Cell Sci. 118:3419-3430) has shown that localization of nesp2G to the ONM is dependent upon an interaction with Sun1. In this study, we confirm and extend these results by demonstrating that both Sun1 and Sun2 contribute to nesp2G localization. Codepletion of both of these proteins in HeLa cells leads to the loss of ONM-associated nesp2G, as does overexpression of the Sun1 lumenal domain. Both treatments result in the expansion of the PNS. These data, together with those of Padmakumar et al. (2005), support a model in which Sun proteins tether nesprins in the ONM via interactions spanning the PNS. In this way, Sun proteins and nesprins form a complex that links the nucleoskeleton and cytoskeleton (the LINC complex).
Figures





References
-
- Apel, E.D., R.M. Lewis, R.M. Grady, and J.R. Sanes. 2000. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275:31986–31995. - PubMed
-
- Broers, J.L., E.A. Peeters, H.J. Kuijpers, J. Endert, C.V. Bouten, C.W. Oomens, F.P. Baaijens, and F.C. Ramaekers. 2004. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 13:2567–2580. - PubMed
-
- Burke, B., and C.L. Stewart. 2002. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 3:575–585. - PubMed
-
- Burnette, W.N. 1981. ‘Western blotting’: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195–203. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

