Genomewide scan and fine-mapping linkage studies in four European samples with bipolar affective disorder suggest a new susceptibility locus on chromosome 1p35-p36 and provides further evidence of loci on chromosome 4q31 and 6q24
- PMID: 16380920
- PMCID: PMC1285167
- DOI: 10.1086/498619
Genomewide scan and fine-mapping linkage studies in four European samples with bipolar affective disorder suggest a new susceptibility locus on chromosome 1p35-p36 and provides further evidence of loci on chromosome 4q31 and 6q24
Abstract
We present the findings of a large linkage study of bipolar affective disorder (BPAD) that involved genomewide analysis of 52 families (448 genotyped individuals) of Spanish, Romany, and Bulgarian descent and further fine mapping of the 1p34-p36, 4q28-q31, and 6q15-q24 regions. An additional sample of 56 German families (280 individuals) was included for this fine-mapping step. The highest nonparametric linkage scores obtained in the fine mapping were 5.49 for 4q31 and 4.87 for 6q24 in the Romany families and 3.97 for 1p35-p36 in the Spanish sample. MOD-score (LOD scores maximized over genetic model parameters) analysis provided significant evidence of linkage to 4q31 and at least borderline significance for the 1p and 6q regions. On the basis of these results and previous positive research findings, 4q31 and 6q24 should now be considered confirmed BPAD susceptibility loci, and 1p35-p36 is proposed as a new putative locus that requires confirmation in replication studies.
Figures






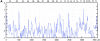








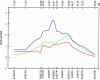




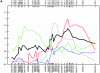



References
Web Resources
-
- deCODE Genetics, http://www.decode.com/ (for information about the genetic map)
-
- NCBI, http://www.ncbi.nih.gov/ (for BPAD)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BPAD) - PubMed
-
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for marker positions and the RefSeq Genes track)
References
-
- American Psychiatric Association (1994) DSM-IV: diagnostic and statistical manual of mental disorders, 4th ed. American Psychiatric Association, Washington, DC
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

