Transcriptional response of steady-state yeast cultures to transient perturbations in carbon source
- PMID: 16381818
- PMCID: PMC1326188
- DOI: 10.1073/pnas.0509978103
Transcriptional response of steady-state yeast cultures to transient perturbations in carbon source
Abstract
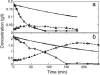
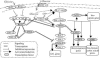
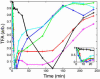
To understand the dynamics of transcriptional response to changing environments, well defined, easily controlled, and short-term perturbation experiments were undertaken. We subjected steady-state cultures of Saccharomyces cerevisiae in chemostats growing on limiting galactose to two different size pulses of glucose, well known to be a preferred carbon source. Although these pulses were not large enough to change growth rates or cell size, approximately 25% of the genes changed their expression at least 2-fold. Using DNA microarrays to estimate mRNA abundance, we found a number of distinguishable patterns of transcriptional response among the many genes whose expression changed. Many of these genes were already known to be regulated by particular transcription factors; we estimated five potentially relevant transcription factor activities from the observed changes in gene expression (i.e., Mig1p, Gal4p, Cat8p, Rgt1p, Adr1p, and Rcs1p). With these estimates, for two regulatory circuits involving interaction among multiple regulators we could generate dynamical models that quantitatively account for the observed transcriptional responses to the transient perturbations.
Figures



References
-
- Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109-126. - PubMed
-
- Ideker, T., Thorsson, V., Ranish, J. A., Christmas, R., Buhler, J., Eng, J. K., Bumgarner, R., Goodlett, D. R., Aebersold, R. & Hood, L. (2001) Science 292, 929-934. - PubMed
-
- Johnston, M. (1999) Trends Genet. 15, 29-33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

