The tmRDB and SRPDB resources
- PMID: 16381838
- PMCID: PMC1347504
- DOI: 10.1093/nar/gkj142
The tmRDB and SRPDB resources
Abstract
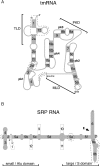
Maintained at the University of Texas Health Science Center at Tyler, Texas, the tmRNA database (tmRDB) is accessible at the URL http://psyche.uthct.edu/dbs/tmRDB/tmRDB.html with mirror sites located at Auburn University, Auburn, Alabama (http://www.ag.auburn.edu/mirror/tmRDB/) and the Royal Veterinary and Agricultural University, Denmark (http://tmrdb.kvl.dk/). The signal recognition particle database (SRPDB) at http://psyche.uthct.edu/dbs/SRPDB/SRPDB.html is mirrored at http://srpdb.kvl.dk/ and the University of Goteborg (http://bio.lundberg.gu.se/dbs/SRPDB/SRPDB.html). The databases assist in investigations of the tmRNP (a ribonucleoprotein complex which liberates stalled bacterial ribosomes) and the SRP (a particle which recognizes signal sequences and directs secretory proteins to cell membranes). The curated tmRNA and SRP RNA alignments consider base pairs supported by comparative sequence analysis. Also shown are alignments of the tmRNA-associated proteins SmpB, ribosomal protein S1, alanyl-tRNA synthetase and Elongation Factor Tu, as well as the SRP proteins SRP9, SRP14, SRP19, SRP21, SRP54 (Ffh), SRP68, SRP72, cpSRP43, Flhf, SRP receptor (alpha) and SRP receptor (beta). All alignments can be easily examined using a new exploratory browser. The databases provide links to high-resolution structures and serve as depositories for structures obtained by molecular modeling.
Figures
References
-
- Karzai A.W., Roche E.D., Sauer R.T. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nature Struct. Biol. 2000;7:449–455. - PubMed
-
- Halic M., Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Biol. 2005;15:116–125. - PubMed
-
- Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. - PubMed
-
- Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. - PubMed
-
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

