LOCATE: a mouse protein subcellular localization database
- PMID: 16381849
- PMCID: PMC1347432
- DOI: 10.1093/nar/gkj069
LOCATE: a mouse protein subcellular localization database
Abstract
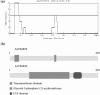
We present here LOCATE, a curated, web-accessible database that houses data describing the membrane organization and subcellular localization of proteins from the FANTOM3 Isoform Protein Sequence set. Membrane organization is predicted by the high-throughput, computational pipeline MemO. The subcellular locations of selected proteins from this set were determined by a high-throughput, immunofluorescence-based assay and by manually reviewing >1700 peer-reviewed publications. LOCATE represents the first effort to catalogue the experimentally verified subcellular location and membrane organization of mammalian proteins using a high-throughput approach and provides localization data for approximately 40% of the mouse proteome. It is available at http://locate.imb.uq.edu.au.
Figures
References
-
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
-
- Nielsen H., Krogh A. In: Sixth International Conference on Intelligent Systems for Molecular Biology. Glasgow J., editor. Vol. 1. AAAI Press; 1998. pp. 122–130. - PubMed
-
- Tusnady G.E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. - PubMed
-
- Krogh A., Larsson B., vonHeijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

