BIOZON: a hub of heterogeneous biological data
- PMID: 16381854
- PMCID: PMC1347515
- DOI: 10.1093/nar/gkj153
BIOZON: a hub of heterogeneous biological data
Abstract
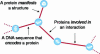
Biological entities are strongly related and mutually dependent on each other. Therefore, there is a growing need to corroborate and integrate data from different resources and aspects of biological systems in order to analyze them effectively. Biozon is a unified biological database that integrates heterogeneous data types such as proteins, structures, domain families, protein-protein interactions and cellular pathways, and establishes the relationships between them. All data are integrated on to a single graph schema centered around the non-redundant set of biological objects that are shared by each source. This integration results in a highly connected graph structure that provides a more complete picture of the known context of a given object that cannot be determined from any one source. Currently, Biozon integrates roughly 2 million protein sequences, 42 million DNA or RNA sequences, 32,000 protein structures, 150,000 interactions and more from sources such as GenBank, UniProt, Protein Data Bank (PDB) and BIND. Biozon augments source data with locally derived data such as 5 billion pairwise protein alignments and 8 million structural alignments. The user may form complex cross-type queries on the graph structure, add similarity relations to form fuzzy queries and rank the results based on analysis of the edge structure similar to Google PageRank, online at Biozon.org.
Figures