HUMHOT: a database of human meiotic recombination hot spots
- PMID: 16381857
- PMCID: PMC1347372
- DOI: 10.1093/nar/gkj009
HUMHOT: a database of human meiotic recombination hot spots
Abstract
Meiotic recombination occurs preferentially at certain regions in the genome referred to as hot spots. The number of hot spots known in humans has increased manifold in recent years. The identification of these hot spots in humans is of great interest to population and medical geneticists since they influence the structure of Linkage Disequilibrium and Haplotype blocks in human populations, whose patterns have applications in mapping disease genes. HUMHOT is a web-based database of Human Meiotic Recombination Hot Spots. The database comprises DNA sequences corresponding to the hot spot regions from the literature that have been mapped to a high resolution (<4 kb) in humans. It also provides flanking sequence information for the hot spot region along with references describing the hot spot. The database can be queried based on hot spot identity, chromosome position or by homology to user-defined sequences. It is also updated with new hot spot sequences as they are discovered and provides hyperlinks to commonly used tools for estimating recombination rates, performing genetic analysis and new advances in our understanding of meiotic hot spots. Public access to the HUMHOT database is available at http://www.jncasr.ac.in/humhot.
Figures
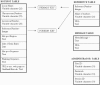
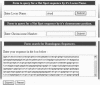

Similar articles
-
Molecular features of meiotic recombination hot spots.Bioessays. 2006 Jan;28(1):45-56. doi: 10.1002/bies.20349. Bioessays. 2006. PMID: 16369948 Review.
-
Human recombination hot spots hidden in regions of strong marker association.Nat Genet. 2005 Jun;37(6):601-6. doi: 10.1038/ng1565. Epub 2005 May 8. Nat Genet. 2005. PMID: 15880103
-
Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex.Nat Genet. 2001 Oct;29(2):217-22. doi: 10.1038/ng1001-217. Nat Genet. 2001. PMID: 11586303
-
Allele-dependent recombination frequency: homology requirement in meiotic recombination at the hot spot in the mouse major histocompatibility complex.Genomics. 1995 May 20;27(2):298-305. doi: 10.1006/geno.1995.1046. Genomics. 1995. PMID: 7557996
-
Meiotic recombination hot spots and cold spots.Nat Rev Genet. 2001 May;2(5):360-9. doi: 10.1038/35072078. Nat Rev Genet. 2001. PMID: 11331902 Review.
Cited by
-
Distributions of single nucleotide polymorphisms in differential chromosome segments of congenic resistant strains that define minor histocompatibility antigens.Immunogenetics. 2007 Aug;59(8):631-9. doi: 10.1007/s00251-007-0231-9. Epub 2007 May 31. Immunogenetics. 2007. PMID: 17541577
-
Joint Prediction of the Effective Population Size and the Rate of Fixation of Deleterious Mutations.Genetics. 2016 Nov;204(3):1267-1279. doi: 10.1534/genetics.116.188250. Epub 2016 Sep 26. Genetics. 2016. PMID: 27672094 Free PMC article.
-
A structured coalescent model reveals deep ancestral structure shared by all modern humans.Nat Genet. 2025 Apr;57(4):856-864. doi: 10.1038/s41588-025-02117-1. Epub 2025 Mar 18. Nat Genet. 2025. PMID: 40102687 Free PMC article.
-
LDSplitDB: a database for studies of meiotic recombination hotspots in MHC using human genomic data.BMC Med Genomics. 2018 Apr 20;11(Suppl 2):27. doi: 10.1186/s12920-018-0351-0. BMC Med Genomics. 2018. PMID: 29697370 Free PMC article.
-
Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22).Am J Hum Genet. 2006 Sep;79(3):524-38. doi: 10.1086/507652. Epub 2006 Aug 1. Am J Hum Genet. 2006. PMID: 16909390 Free PMC article.
References
-
- Sun H., Treco D., Schultes N.P., Szostak J.W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. - PubMed
-
- Keeney S., Giroux C.N., Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. - PubMed
-
- Kauppi L., Jeffreys A.J., Keeney S. Where the crossovers are: recombination distributions in mammals. Nature Rev. Genet. 2004;5:413–424. - PubMed
-
- Smith G.R., Amundsen S.K., Dabert P., Taylor A.F. The initiation and control of homologous recombination in E.coli. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1995;347:13–20. - PubMed
-
- Hubert R., MacDonald M., Gusella J., Arnheim N. High resolution localization of recombination hot spots using sperm typing. Nature Genet. 1994;7:420–424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

