EMAGE: a spatial database of gene expression patterns during mouse embryo development
- PMID: 16381949
- PMCID: PMC1347369
- DOI: 10.1093/nar/gkj006
EMAGE: a spatial database of gene expression patterns during mouse embryo development
Abstract
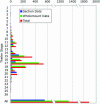
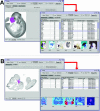
EMAGE (http://genex.hgu.mrc.ac.uk/Emage/database) is a freely available, curated database of gene expression patterns generated by in situ techniques in the developing mouse embryo. It is unique in that it contains standardized spatial representations of the sites of gene expression for each gene, denoted against a set of virtual reference embryo models. As such, the data can be interrogated in a novel and abstract manner by using space to define a query. Accompanying the spatial representations of gene expression patterns are text descriptions of the sites of expression, which also allows searching of the data by more conventional text-based methods.
Figures
References
-
- Baldock R.A., Bard J.B., Burger A., Burton N., Christiansen J., Feng G., Hill B., Houghton D., Kaufman M., Rao J., et al. EMAP and EMAGE: a framework for understanding spatially organized data. Neuroinformatics. 2003;1:309–325. - PubMed
-
- Kaufman M. The Atlas of Mouse Development. London: Academic Press; 1992.
-
- Theiler K. The House Mouse: Atlas of Embryonic Development. NY: Springer-Verlag; 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

