Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast
- PMID: 16382152
- PMCID: PMC1346895
- DOI: 10.1128/MCB.26.2.617-629.2006
Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast
Abstract
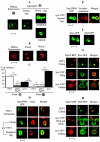
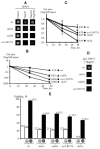
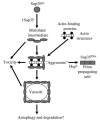
Self-perpetuating protein aggregates transmit prion diseases in mammals and heritable traits in yeast. De novo prion formation can be induced by transient overproduction of the corresponding prion-forming protein or its prion domain. Here, we demonstrate that the yeast prion protein Sup35 interacts with various proteins of the actin cortical cytoskeleton that are involved in endocytosis. Sup35-derived aggregates, generated in the process of prion induction, are associated with the components of the endocytic/vacuolar pathway. Mutational alterations of the cortical actin cytoskeleton decrease aggregation of overproduced Sup35 and de novo prion induction and increase prion-related toxicity in yeast. Deletion of the gene coding for the actin assembly protein Sla2 is lethal in cells containing the prion isoforms of both Sup35 and Rnq1 proteins simultaneously. Our data are consistent with a model in which cytoskeletal structures provide a scaffold for generation of large aggregates, resembling mammalian aggresomes. These aggregates promote prion formation. Moreover, it appears that the actin cytoskeleton also plays a certain role in counteracting the toxicity of the overproduced potentially aggregating proteins.
Figures



References
-
- Allen, K. D., R. D. Wegrzyn, T. A. Chernova, S. Müller, G. P. Newnam, P. A. Winslett, K. B. Wittich, K. D. Wilkinson, and Y. O. Chernoff. 2005. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169:1227-1242. - PMC - PubMed
-
- Ayscough, K. R. 2000. Endocytosis and the development of cell polarity in yeast require a dynamic F-actin cytoskeleton. Curr. Biol. 10:1587-1590. - PubMed
-
- Ayscough, K. R., J. J. Eby, T. Lila, H. Dewar, K. G. Kozminski, and D. G. Drubin. 1999. Sla1p is a functionally modular component of the yeast cortical actin cytoskeleton required for correct localization of both Rho1p-GTPase and Sla2p, a protein with talin homology. Mol. Biol. Cell 10:1061-1075. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
