The rice mitochondrial genomes and their variations
- PMID: 16384910
- PMCID: PMC1361312
- DOI: 10.1104/pp.105.070060
The rice mitochondrial genomes and their variations
Abstract
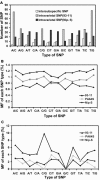
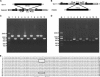
Based on highly redundant and high-quality sequences, we assembled rice (Oryza sativa) mitochondrial genomes for two cultivars, 93-11 (an indica variety) and PA64S (an indica-like variety with maternal origin of japonica), which are paternal and maternal strains of an elite superhybrid rice Liang-You-Pei-Jiu (LYP-9), respectively. Following up with a previous analysis on rice chloroplast genomes, we divided mitochondrial sequence variations into two basic categories, intravarietal and intersubspecific. Intravarietal polymorphisms are variations within mitochondrial genomes of an individual variety. Intersubspecific polymorphisms are variations between subspecies among their major genotypes. In this study, we identified 96 single nucleotide polymorphisms (SNPs), 25 indels, and three segmental sequence variations as intersubspecific polymorphisms. A signature sequence fragment unique to indica varieties was confirmed experimentally and found in two wild rice samples, but absent in japonica varieties. The intersubspecific polymorphism rate for mitochondrial genomes is 0.02% for SNPs and 0.006% for indels, nearly 2.5 and 3 times lower than that of their chloroplast counterparts and 21 and 38 times lower than corresponding rates of the rice nuclear genome, respectively. The intravarietal polymorphism rates among analyzed mitochondrial genomes, such as 93-11 and PA64S, are 1.26% and 1.38% for SNPs and 1.13% and 1.09% for indels, respectively. Based on the total number of SNPs between the two mitochondrial genomes, we estimate that the divergence of indica and japonica mitochondrial genomes occurred approximately 45,000 to 250,000 years ago.
Figures


References
-
- Dai ZY, Zhao BH, Liu XJ (1997) A new medium indica variety with fine quality, high yield and multi-disease resistance. Jiangsu Agricultural Science 1: 13–14
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

