YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport
- PMID: 16385049
- PMCID: PMC1347309
- DOI: 10.1128/JB.188.2.587-598.2006
YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport
Abstract
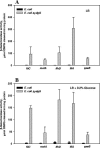
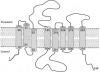
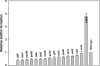
YdgG is an uncharacterized protein that is induced in Escherichia coli biofilms. Here it is shown that deletion of ydgG decreased extracellular and increased intracellular concentrations of autoinducer 2 (AI-2); hence, YdgG enhances transport of AI-2. Consistent with this hypothesis, deletion of ydgG resulted in a 7,000-fold increase in biofilm thickness and 574-fold increase in biomass in flow cells. Also consistent with the hypothesis, deletion of ydgG increased cell motility by increasing transcription of flagellar genes (genes induced by AI-2). By expressing ydgG in trans, the wild-type phenotypes for extracellular AI-2 activity, motility, and biofilm formation were restored. YdgG is also predicted to be a membrane-spanning protein that is conserved in many bacteria, and it influences resistance to several antimicrobials, including crystal violet and streptomycin (this phenotype could also be complemented). Deletion of ydgG also caused 31% of the bacterial chromosome to be differentially expressed in biofilms, as expected, since AI-2 controls hundreds of genes. YdgG was found to negatively modulate expression of flagellum- and motility-related genes, as well as other known products essential for biofilm formation, including operons for type 1 fimbriae, autotransporter protein Ag43, curli production, colanic acid production, and production of polysaccharide adhesin. Eighty genes not previously related to biofilm formation were also identified, including those that encode transport proteins (yihN and yihP), polysialic acid production (gutM and gutQ), CP4-57 prophage functions (yfjR and alpA), methionine biosynthesis (metR), biotin and thiamine biosynthesis (bioF and thiDFH), anaerobic metabolism (focB, hyfACDR, ttdA, and fumB), and proteins with unknown function (ybfG, yceO, yjhQ, and yjbE); 10 of these genes were verified through mutation to decrease biofilm formation by 40% or more (yfjR, bioF, yccW, yjbE, yceO, ttdA, fumB, yjiP, gutQ, and yihR). Hence, it appears YdgG controls the transport of the quorum-sensing signal AI-2, and so we suggest the gene name tqsA.
Figures


References
-
- Baba, T., T. Ara, Y. Okumura, M. I. Hasegawa, Y. Takai, M. Baba, T. Oshima, M. Tomita, B. Wanner, and H. Mori. Unpublished data.
-
- Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. - PubMed
-
- Beloin, C., and J.-M. Ghigo. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 13:16-19. - PubMed
-
- Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. J. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J.-M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

