Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans
- PMID: 16385060
- PMCID: PMC1347274
- DOI: 10.1128/JB.188.2.702-710.2006
Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans
Abstract
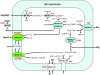
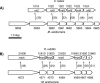
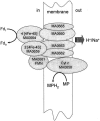
A liquid chromatography-hybrid linear ion trap-Fourier transform ion cyclotron resonance mass spectrometry approach was used to determine the differential abundance of proteins in acetate-grown cells compared to that of proteins in methanol-grown cells of the marine isolate Methanosarcina acetivorans metabolically labeled with 14N versus 15N. The 246 differentially abundant proteins in M. acetivorans were compared with the previously reported 240 differentially expressed genes of the freshwater isolate Methanosarcina mazei determined by transcriptional profiling of acetate-grown cells compared to methanol-grown cells. Profound differences were revealed for proteins involved in electron transport and energy conservation. Compared to methanol-grown cells, acetate-grown M. acetivorans synthesized greater amounts of subunits encoded in an eight-gene transcriptional unit homologous to operons encoding the ion-translocating Rnf electron transport complex previously characterized from the Bacteria domain. Combined with sequence and physiological analyses, these results suggest that M. acetivorans replaces the H2-evolving Ech hydrogenase complex of freshwater Methanosarcina species with the Rnf complex, which generates a transmembrane ion gradient for ATP synthesis. Compared to methanol-grown cells, acetate-grown M. acetivorans synthesized a greater abundance of proteins encoded in a seven-gene transcriptional unit annotated for the Mrp complex previously reported to function as a sodium/proton antiporter in the Bacteria domain. The differences reported here between M. acetivorans and M. mazei can be attributed to an adaptation of M. acetivorans to the marine environment.
Figures



References
-
- Andreev, V. P., Z. Zang, T. Rejtar, S. Pillai, L. Li, Y. Wang, S.-W. Wu, and B. L. Karger. 2005. A new algorithm for quantitation of LC-MS proteomic data. Proceedings of the 53rd ASMS Conference on Mass Spectrometry and Allied Topics. The American Society for Mass Spectrometry, San Antonio, Tex.
-
- Barquera, B., C. C. Hase, and R. B. Gennis. 2001. Expression and mutagenesis of the NqrC subunit of the NQR respiratory Na+ pump from Vibrio cholerae with covalently attached FMN. FEBS Lett. 492:45-49. - PubMed
-
- Beattie, P., K. Tan, R. M. Bourne, D. Leach, P. R. Rich, and F. B. Ward. 1994. Cloning and sequencing of four structural genes for the Na+-translocating NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett. 356:333-338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

