Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2
- PMID: 16385450
- PMCID: PMC1380224
- DOI: 10.1086/498853
Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2
Abstract
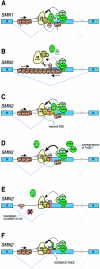
Spinal muscular atrophy is a neurodegenerative disorder caused by the deletion or mutation of the survival-of-motor-neuron gene, SMN1. An SMN1 paralog, SMN2, differs by a C-->T transition in exon 7 that causes substantial skipping of this exon, such that SMN2 expresses only low levels of functional protein. A better understanding of SMN splicing mechanisms should facilitate the development of drugs that increase survival motor neuron (SMN) protein levels by improving SMN2 exon 7 inclusion. In addition, exonic mutations that cause defective splicing give rise to many genetic diseases, and the SMN1/2 system is a useful paradigm for understanding exon-identity determinants and alternative-splicing mechanisms. Skipping of SMN2 exon 7 was previously attributed either to the loss of an SF2/ASF-dependent exonic splicing enhancer or to the creation of an hnRNP A/B-dependent exonic splicing silencer, as a result of the C-->T transition. We report the extensive testing of the enhancer-loss and silencer-gain models by mutagenesis, RNA interference, overexpression, RNA splicing, and RNA-protein interaction experiments. Our results support the enhancer-loss model but also demonstrate that hnRNP A/B proteins antagonize SF2/ASF-dependent ESE activity and promote exon 7 skipping by a mechanism that is independent of the C-->T transition and is, therefore, common to both SMN1 and SMN2. Our findings explain the basis of defective SMN2 splicing, illustrate the fine balance between positive and negative determinants of exon identity and alternative splicing, and underscore the importance of antagonistic splicing factors and exonic elements in a disease context.
Figures







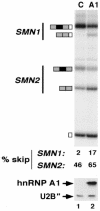
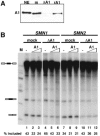
References
Web Resources
-
- Entrez Gene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene/ (for SMN1, SMN2, and CD44)
-
- ESEfinder 2.0, http://rulai.cshl.edu/tools/ESE/ (for identifying putative SR protein–dependent ESEs and calculating motif scores)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SMA types I, II, and III)
-
- Pictogram, http://genes.mit.edu/pictogram.html/ (for pictogram representations of position weight matrices)
-
- RESCUE-ESE, http://genes.mit.edu/burgelab/rescue-ese/ (for identifying candidate ESEs)
References
-
- Bourgeois CF, Lejeune F, Stévenin J (2004) Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol 78:37–88 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

